AIDO.StructureDecoder
AIDO.StructureDecoder is the decoder-only component of AIDO.StructureTokenizer for tokenization of protein structures.
Model Description
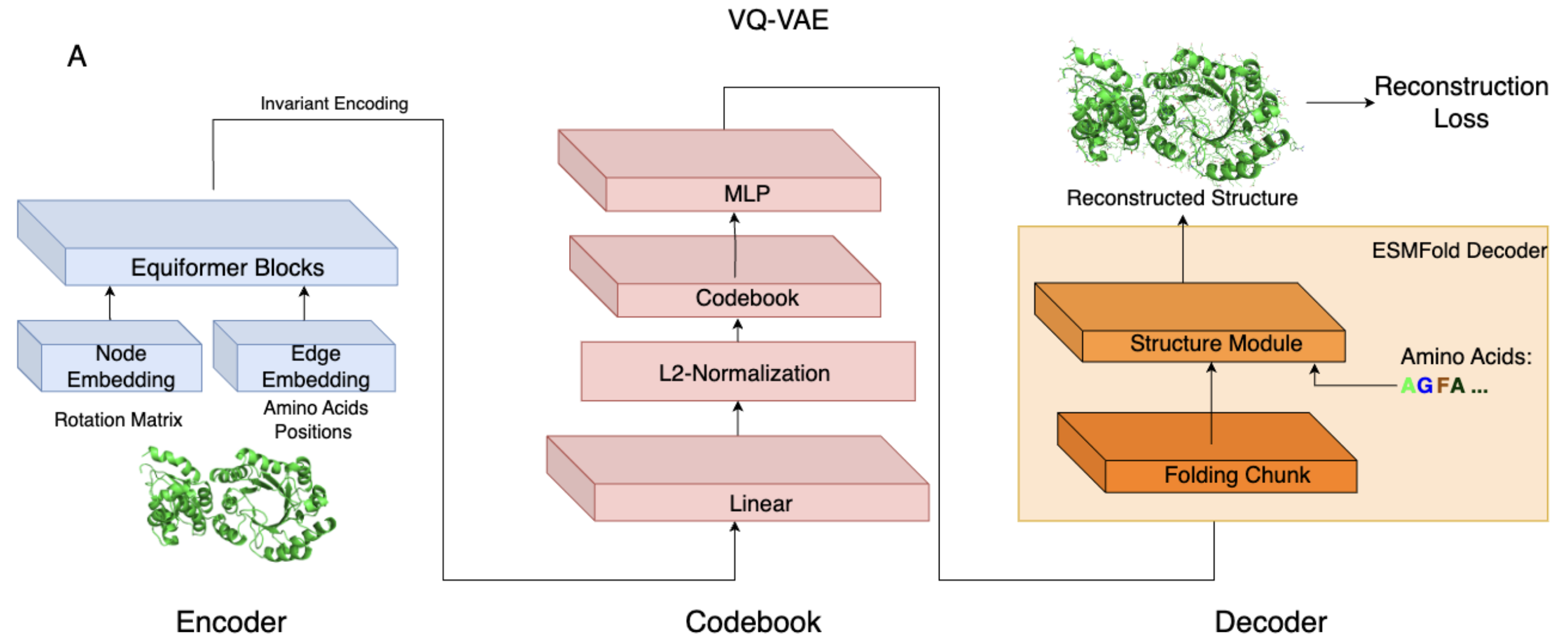
AIDO.StructureTokenizer is built on a Vector Quantized Variational Autoencoder (VQ-VAE) architecture with the following components:
- Equivariant Encoder (6M): Encodes backbone structures into a latent space that maintains rotational and translational symmetries using the Equiformer architecture.
- Discrete Codebook: Maps continuous latent vectors into 512 discrete structural tokens.
- Invariant Decoder (300M): Reconstructs full 3D structures, including side chains, from the structural tokens using an architecture adapted from ESMFold.
This model strikes a balance between reconstruction fidelity and structural locality, optimizing its suitability for downstream tasks such as structure prediction, homology detection, and multimodal protein modeling.
Key Features
- Encoding Structures into Tokens (See genbio-ai/AIDO.StructureEncoder)
- Decoding Tokens into Structures (See below)
- Reconstructing Structures (See genbio-ai/AIDO.StructureTokenizer)
- Structure Prediction (See this section in genbio-ai/AIDO.Protein2StructureToken-16B)
How to Use
Please see experiments/AIDO.StructureTokenizer in Model Generator for more details.
Setup
Install Model Generator
Decoding Structure Tokens from AIDO.StructureEncoder
If you have run the encoding task in genbio-ai/AIDO.StructureEncoder with the default encode.yaml, the default decode.yaml configuration file is already set up to decode the encoded tokens. You don't need to change anything in the configuration file. You can directly run the decoding task using the following command:
CUDA_VISIBLE_DEVICES=0 mgen predict --config=experiments/AIDO.StructureTokenizer/decode.yaml
Decoding Customized Structure Tokens
To decode protein structures, you will need the structure tokens in .pt format and a corresponding codebook file (codebook.pt). For ease of use, we recommend preparing the structure tokens in TSV format and then converting them to .pt format using the provided script.
The TSV file should include the following columns (an example file is available at experiments/AIDO.StructureTokenizer/decode_example_input.tsv):
uid: A unique identifier for the protein sequence.sequences: The amino acid sequence (e.g., "LRTPTT").predictions: The structure tokens to be decoded, provided as a list (e.g., "[164, 287, 119, ...]"). The list length must match the length of the amino acid sequence.
After preparing the TSV file, you need to convert the TSV file to the .pt format using the following command:
python experiments/AIDO.StructureTokenizer/struct_token_format_conversion.py your_tsv_file.tsv your_output_pt_file.pt
You also need to prepare a codebook file (codebook.pt) that contains the embedding of each token. The codebook could be extracted using this command:
python experiments/AIDO.StructureTokenizer/extract_structure_tokenizer_codebook.py --output_path your_output_codebook.pt
Then you need to update the struct_tokens_path and codebook_path in the decode.yaml configuration file to point to your structure tokens and codebook file. Alternatively, you can override these parameters when running the command:
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/decode.yaml \
--data.init_args.config.struct_tokens_datasets_configs.name="your_dataset_name" \
--data.init_args.config.struct_tokens_datasets_configs.struct_tokens_path="your_structure_tokens.pt" \
--data.init_args.config.struct_tokens_datasets_configs.codebook_path="your_codebook.pt" \
--trainer.callbacks.dict_kwargs.dirpath="your_output_dir"
Input:
- The encoded tokens saved in
.ptformat. - The codebook file (
codebook.pt) that contains the embedding of each token.
Output:
- The decoded protein structures will be saved in the output directory specified in the configuration file. By default it is saved in
logs/protstruct_decode/. - The decoded structures are saved as PDB files.
Notes:
- Decoding the structures could take a long time even when using a GPU.
- Currently, this function only supports single GPU inference due to the file saving mechanism. We plan to support multi-GPU inference in the future.
- Currently, we don't support specifying the residue index in TSV format. If you need to specify the residue index, you need to modify the
struct_token_format_conversion.pyscript to include the residue index in the TSV file (we may support this feature in the future), or you could provide the.ptfile directly with the desired residue index.
Citation
Please cite AIDO.StructureTokenizer using the following BibTex code:
@inproceedings{zhang_balancing_2024,
title = {Balancing Locality and Reconstruction in Protein Structure Tokenizer},
url = {https://www.biorxiv.org/content/10.1101/2024.12.02.626366v2},
doi = {10.1101/2024.12.02.626366},
publisher = {bioRxiv},
author = {Zhang, Jiayou and Meynard-Piganeau, Barthelemy and Gong, James and Cheng, Xingyi and Luo, Yingtao and Ly, Hugo and Song, Le and Xing, Eric},
year = {2024},
booktitle={NeurIPS 2024 Workshop on Machine Learning in Structural Biology (MLSB)},
}
- Downloads last month
- 9