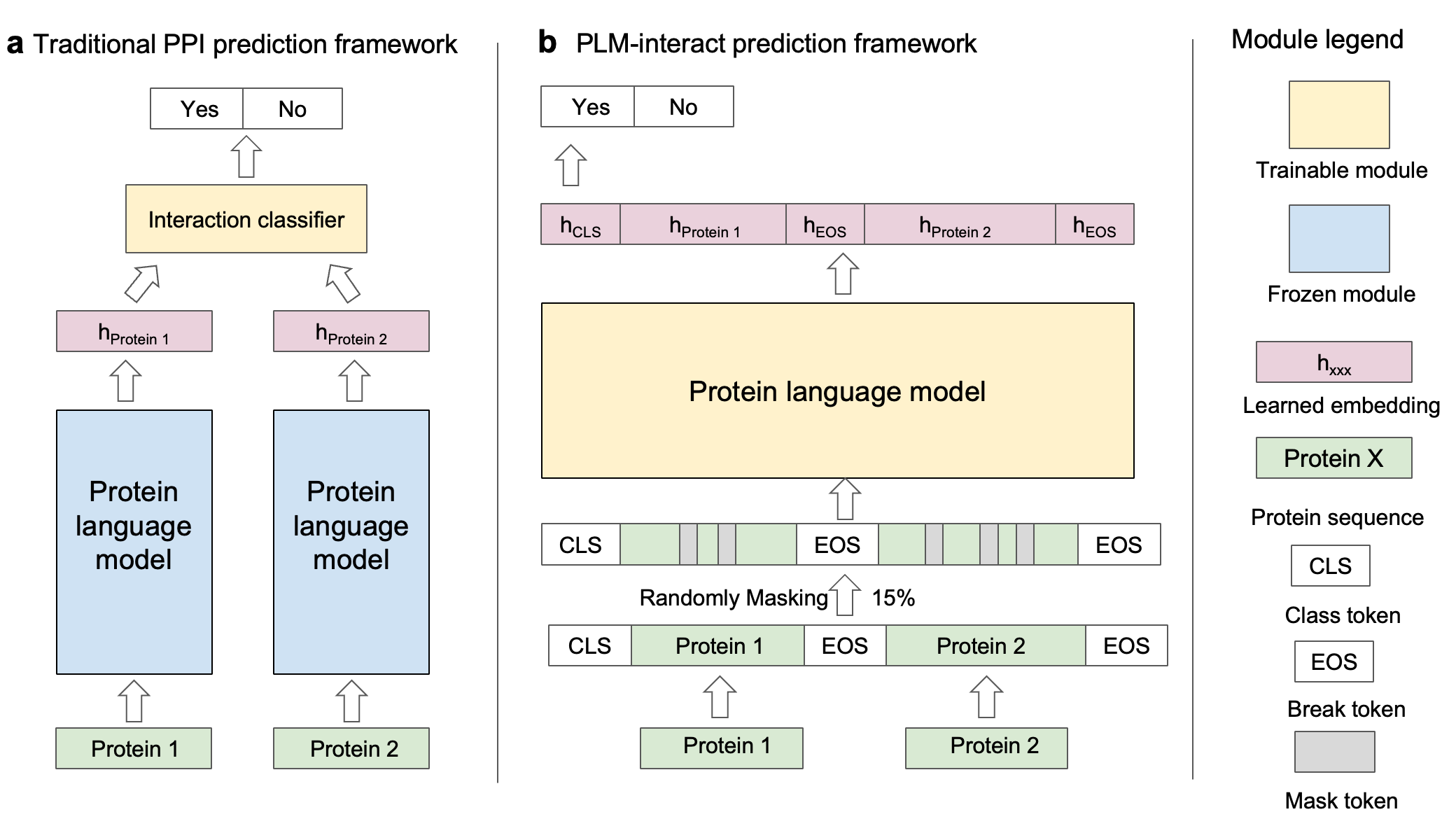
PLM-interact model
PLM-interact: extending protein language models to predict protein-protein interactions The preprint is available at PLM-interact and the code see github link
This model is trained on human PPIs from STRING V12. For the PPI preprocessing details, see Methods in the preprint.
Model description
PLM-interact, goes beyond a single protein, jointly encoding protein pairs to learn their relationships, analogous to the next-sentence prediction task from natural language processing. This approach provides a significant improvement in performance: Trained on human-human PPIs, PLM-interact predicts mouse, fly, worm, E. coli and yeast PPIs, with 16-28% improvements in AUPR compared with state-of-the-art PPI models. Additionally, it can detect changes that disrupt or cause PPIs and be applied to virus-host PPI prediction.
An example to predict interaction probability between proteins
import torch
import torch.nn as nn
from transformers import AutoModel,AutoModelForMaskedLM,AutoTokenizer
import os
import torch.nn.functional as F
class PLMinteract(nn.Module):
def __init__(self,model_name,num_labels,embedding_size):
super(PLMinteract,self).__init__()
self.esm_mask = AutoModelForMaskedLM.from_pretrained(model_name)
self.embedding_size=embedding_size
self.classifier = nn.Linear(embedding_size,1) # embedding_size
self.num_labels=num_labels
def forward_test(self,features):
embedding_output = self.esm_mask.base_model(**features, return_dict=True)
embedding=embedding_output.last_hidden_state[:,0,:] #cls token
embedding = F.relu(embedding)
logits = self.classifier(embedding)
logits=logits.view(-1)
probability = torch.sigmoid(logits)
return probability
# folder_huggingface_download : the download model from huggingface, such as "danliu1226/PLM-interact-650M-humanV11"
# model_name: the ESM2 model that PLM-interact trained
# embedding_size: the embedding size of ESM2 model
folder_huggingface_download='download_huggingface_folder/'
model_name= 'facebook/esm2_t33_650M_UR50D'
embedding_size =1280
protein1 ="EGCVSNLMVCNLAYSGKLEELKESILADKSLATRTDQDSRTALHWACSAGHTEIVEFLLQLGVPVNDKDDAGWSPLHIAASAGRDEIVKALLGKGAQVNAVNQNGCTPLHYAASKNRHEIAVMLLEGGANPDAKDHYEATAMHRAAAKGNLKMIHILLYYKASTNIQDTEGNTPLHLACDEERVEEAKLLVSQGASIYIENKEEKTPLQVAKGGLGLILKRMVEG"
protein2= "MGQSQSGGHGPGGGKKDDKDKKKKYEPPVPTRVGKKKKKTKGPDAASKLPLVTPHTQCRLKLLKLERIKDYLLMEEEFIRNQEQMKPLEEKQEEERSKVDDLRGTPMSVGTLEEIIDDNHAIVSTSVGSEHYVSILSFVDKDLLEPGCSVLLNHKVHAVIGVLMDDTDPLVTVMKVEKAPQETYADIGGLDNQIQEIKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELIQKYLGDGPKLVRELFRVAEEHAPSIVFIDEIDAIGTKRYDSNSGGEREIQRTMLELLNQLDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKKRIFQIHTSRMTLADDVTLDDLIMAKDDLSGADIKAICTEAGLMALRERRMKVTNEDFKKSKENVLYKKQEGTPEGLYL"
DEVICE = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
tokenizer = AutoTokenizer.from_pretrained(model_name)
PLMinter= PLMinteract(model_name, 1, embedding_size)
load_model = torch.load(f"{folder_huggingface_download}pytorch_model.bin")
PLMinter.load_state_dict(load_model)
texts=[protein1, protein2]
tokenized = tokenizer(*texts, padding=True, truncation='longest_first', return_tensors="pt", max_length=1603)
tokenized = tokenized.to(DEVICE)
PLMinter.eval()
PLMinter.to(DEVICE)
with torch.no_grad():
probability = PLMinter.forward_test(tokenized)
print(probability.item())
Training dataset
This model checkpoint is trained on the benchmarking human PPIs from https://d-script.readthedocs.io/en/stable/data.html
- Downloads last month
- 3