MedConvNeXt: Optimized Skin Disease Classification
📌 Introduction
MedConvNeXt is a deep learning model based on ConvNeXt, optimized for skin disease classification using PyTorch Lightning. The model leverages hyperparameter tuning via Optuna to enhance its performance over multiple training iterations.
📂 Dataset
The dataset consists of images of various skin diseases, structured as follows:
SkinDisease/
train/
class_1/
class_2/
...
test/
class_1/
class_2/
...
Data augmentation techniques such as AutoAugment, horizontal flipping, rotation, color jittering, and random erasing were applied to improve model generalization.
⚙️ Model Architecture
- Base Model: ConvNeXt-Base (pretrained on ImageNet)
- Optimizer: AdamW with CosineAnnealingLR scheduler
- Loss Function: CrossEntropyLoss / Focal Loss (for class imbalance handling)
- Evaluation Metrics: Accuracy, Precision, Recall, and F1-score
- Hyperparameter Optimization: Optuna (10 trials, 5 epochs per trial)
📊 Training Process
The model was trained using PyTorch Lightning with automatic logging to TensorBoard for real-time monitoring. The best hyperparameters were selected using Optuna, and the final model was trained over 23 epochs.
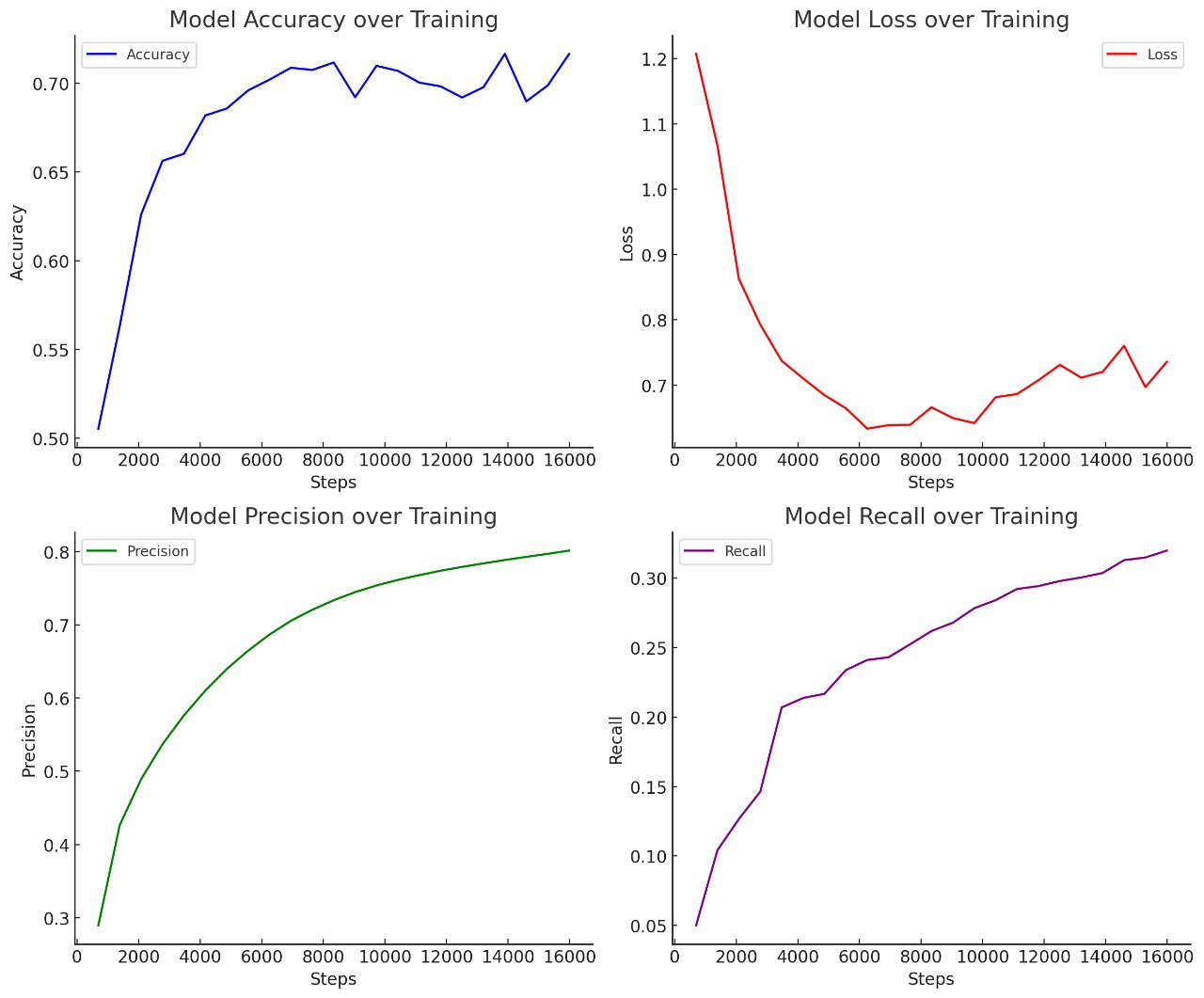
🚀 Results
Below are key performance graphs from TensorBoard:
- Accuracy & Precision improved with hyperparameter tuning
- Training loss consistently decreased, showing model convergence
🔗 How to Use
To load and use the model:
import torch
from torchvision import transforms
from PIL import Image
# Load the model
model = torch.jit.load("skinconvnext_scripted.pt")
model.eval()
# Define image transformation
transform = transforms.Compose([
transforms.Resize((224, 224)),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
# Predict a sample image
image = Image.open("sample.jpg").convert("RGB")
image_tensor = transform(image).unsqueeze(0)
output = model(image_tensor)
predicted_class = torch.argmax(output, dim=1).item()
print("Predicted Class:", predicted_class)
📌 Future Work
- Clinical validation on real-world medical datasets
- Model interpretability via Grad-CAM or SHAP
- Deployment optimization using ONNX and TensorRT
📝 License
This project is intended for research and educational purposes only. For clinical use, further validation is required.
Hugging Face Space: [https://huggingface.co/spaces/Eraly-ml/Skin-AI]
Author: [Eraly Gainulla]
My telegram @eralyf
Model tree for Eraly-ml/Skin-AI
Base model
facebook/convnextv2-tiny-1k-224