Spaces:
Sleeping
Sleeping
Update app.py
Browse files
app.py
CHANGED
|
@@ -417,7 +417,7 @@ with gr.Blocks(theme='ParityError/Interstellar') as demo:
|
|
| 417 |
with gr.Column(min_width=500):
|
| 418 |
gr.Markdown(f"""
|
| 419 |
## How does it work?\n
|
| 420 |
-
|
| 421 |
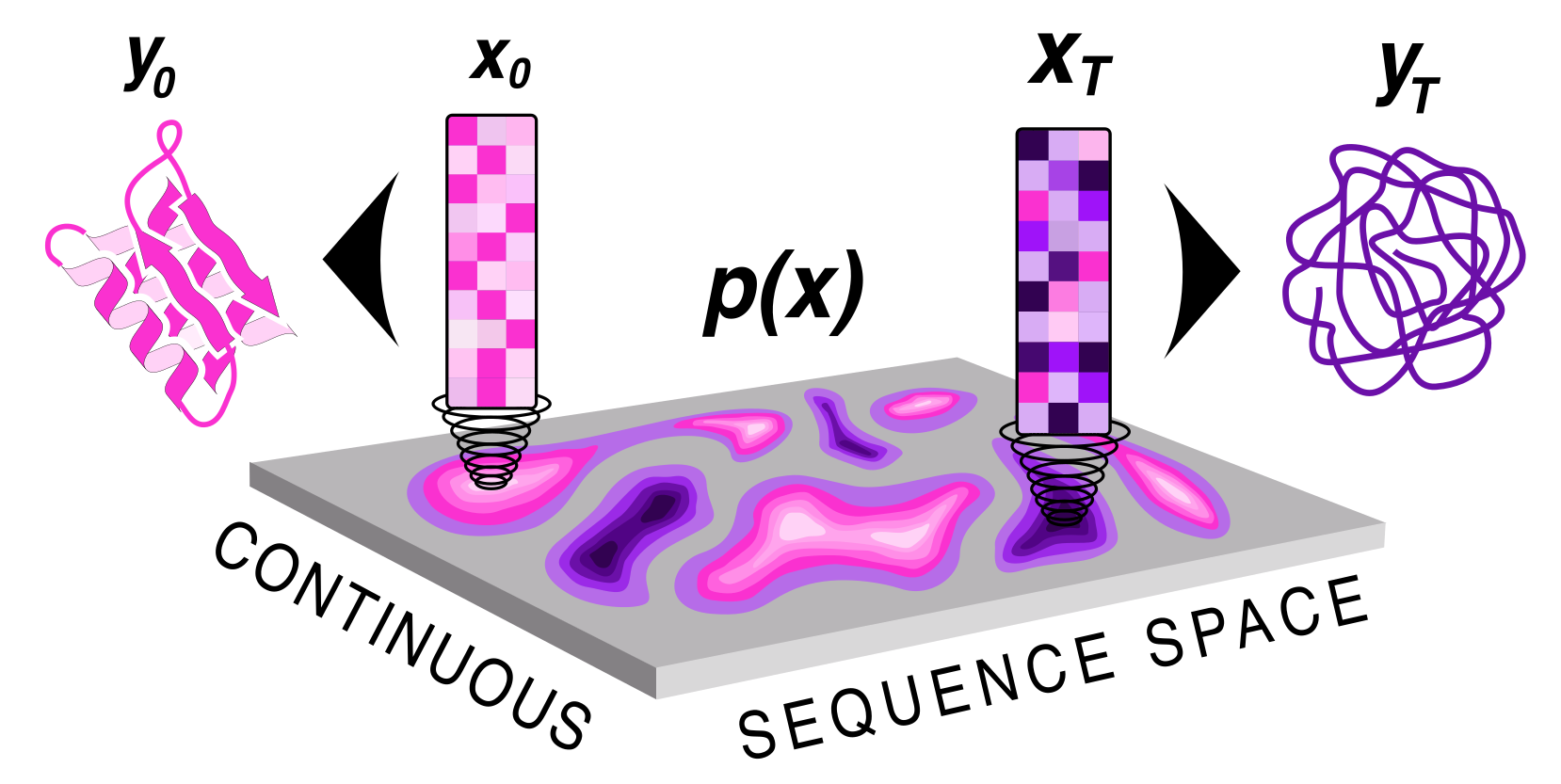
Protein sequence and structure co-generation is a long outstanding problem in the field of protein design. By implementing [ddpm](https://arxiv.org/abs/2006.11239) style diffusion over protein seqeuence space we generate protein sequence and structure pairs. Starting with [RoseTTAFold](https://www.science.org/doi/10.1126/science.abj8754), a protein structure prediction network, we finetuned it to predict sequence and structure given a partially noised sequence. By applying losses to both the predicted sequence and structure the model is forced to generate meaningful pairs. Diffusing in sequence space makes it easy to implement potentials to guide the diffusive process toward particular amino acid composition, net charge, and more! Furthermore, you can sample proteins from a family of sequences or even train a small sequence to function classifier to guide generation toward desired sequences.
|
| 422 |
|
| 423 |
|
|
|
|
| 417 |
with gr.Column(min_width=500):
|
| 418 |
gr.Markdown(f"""
|
| 419 |
## How does it work?\n
|
| 420 |
+
!!! [PAPER](https://www.nature.com/articles/s41587-024-02395-w) !!!
|
| 421 |
Protein sequence and structure co-generation is a long outstanding problem in the field of protein design. By implementing [ddpm](https://arxiv.org/abs/2006.11239) style diffusion over protein seqeuence space we generate protein sequence and structure pairs. Starting with [RoseTTAFold](https://www.science.org/doi/10.1126/science.abj8754), a protein structure prediction network, we finetuned it to predict sequence and structure given a partially noised sequence. By applying losses to both the predicted sequence and structure the model is forced to generate meaningful pairs. Diffusing in sequence space makes it easy to implement potentials to guide the diffusive process toward particular amino acid composition, net charge, and more! Furthermore, you can sample proteins from a family of sequences or even train a small sequence to function classifier to guide generation toward desired sequences.
|
| 422 |

|
| 423 |
|