Spaces:
Sleeping
Sleeping
| #!/usr/bin/env python | |
| # coding: utf-8 | |
| # # Celltype annotation migration(mapping) with TOSICA | |
| # | |
| # We know that when all samples cannot be obtained at the same time, it would be desirable to classify the cell types on the first batch of data and use them to annotate the data obtained later or to be obtained in the future with the same standard, without the need to processing and mapping them together again. | |
| # | |
| # So migration(mapping) the reference cell annotation is necessary. This tutorial focuses on how to migration(mapping) the cell annotation from reference scRNA-seq atlas to new scRNA-seq data. | |
| # | |
| # Paper: [Transformer for one stop interpretable cell type annotation](https://www.nature.com/articles/s41467-023-35923-4) | |
| # | |
| # Code: https://github.com/JackieHanLab/TOSICA | |
| # | |
| # Colab_Reproducibility:https://colab.research.google.com/drive/1BjPEG-kLAgicP8iQvtVtpzzbIOmk1X23?usp=sharing | |
| # | |
| # 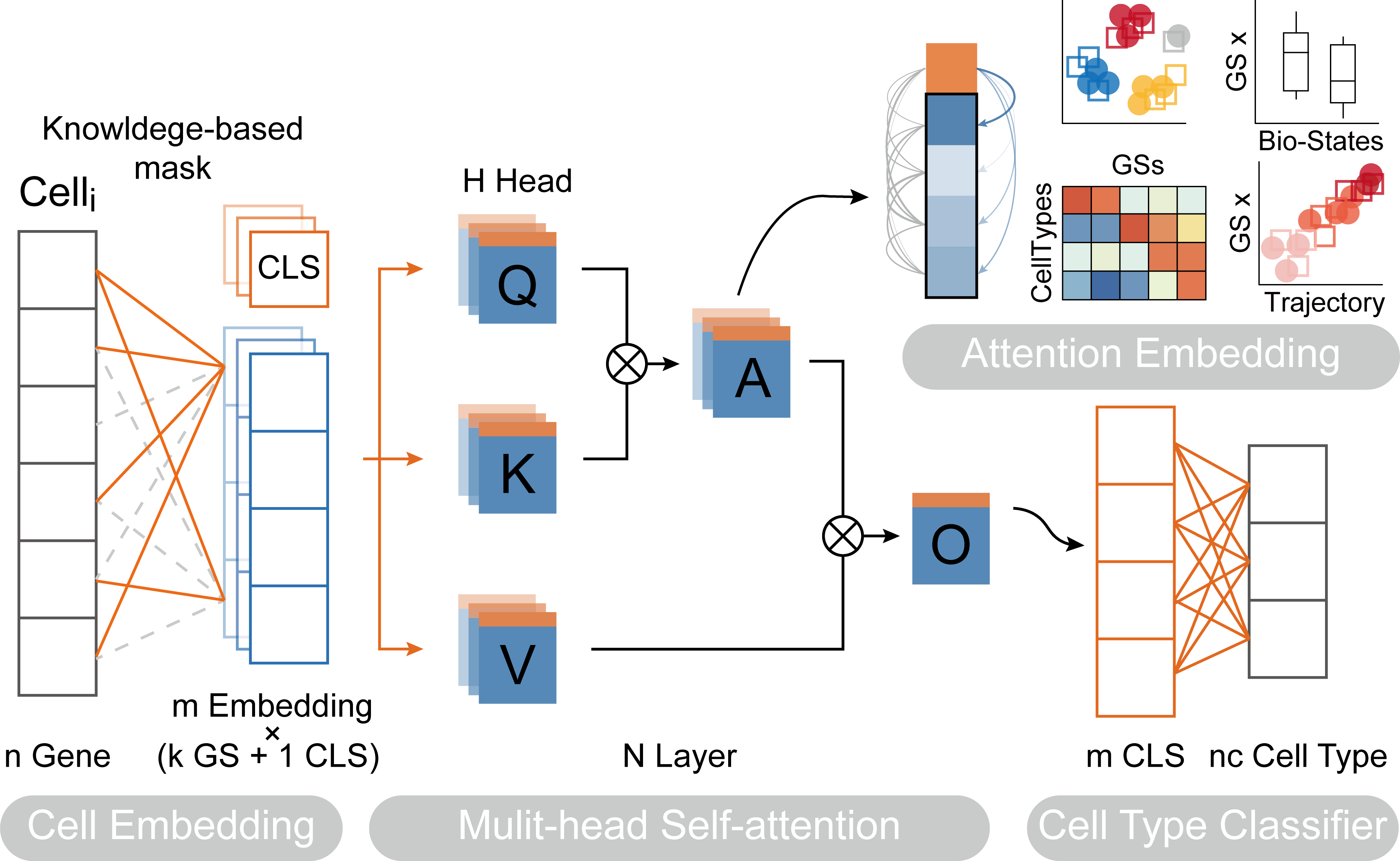 | |
| # | |
| # In[1]: | |
| import omicverse as ov | |
| import scanpy as sc | |
| ov.utils.ov_plot_set() | |
| # ## Loading data | |
| # | |
| # + `demo_train.h5ad` : Braon(GSE84133) and Muraro(GSE85241) | |
| # | |
| # + `demo_test.h5ad` : xin(GSE81608), segerstolpe(E-MTAB-5061) and Lawlor(GSE86473) | |
| # | |
| # They can be downloaded at https://figshare.com/projects/TOSICA_demo/158489. | |
| # In[2]: | |
| ref_adata = sc.read('demo_train.h5ad') | |
| ref_adata = ref_adata[:,ref_adata.var_names] | |
| print(ref_adata) | |
| print(ref_adata.obs.Celltype.value_counts()) | |
| # In[4]: | |
| query_adata = sc.read('demo_test.h5ad') | |
| query_adata = query_adata[:,ref_adata.var_names] | |
| print(query_adata) | |
| print(query_adata.obs.Celltype.value_counts()) | |
| # We need to select the same gene training and predicting the celltype | |
| # In[ ]: | |
| ref_adata.var_names_make_unique() | |
| query_adata.var_names_make_unique() | |
| ret_gene=list(set(query_adata.var_names) & set(ref_adata.var_names)) | |
| len(ret_gene) | |
| # In[ ]: | |
| query_adata=query_adata[:,ret_gene] | |
| ref_adata=ref_adata[:,ret_gene] | |
| # In[5]: | |
| print(f"The max of ref_adata is {ref_adata.X.max()}, query_data is {query_adata.X.max()}",) | |
| # By comparing the maximum values of the two data, we can see that the data has been normalised `sc.pp.normalize_total` and logarithmised `sc.pp.log1p`. The same treatment is applied to the data when we use our own data for analysis. | |
| # | |
| # ## Download Genesets | |
| # | |
| # Here, we need to download the genesets as pathway at first. You can use `ov.utils.download_tosica_gmt()` to download automatically or manual download from: | |
| # | |
| # - 'GO_bp':'https://figshare.com/ndownloader/files/41460072', | |
| # - 'TF':'https://figshare.com/ndownloader/files/41460066', | |
| # - 'reactome':'https://figshare.com/ndownloader/files/41460051', | |
| # - 'm_GO_bp':'https://figshare.com/ndownloader/files/41460060', | |
| # - 'm_TF':'https://figshare.com/ndownloader/files/41460057', | |
| # - 'm_reactome':'https://figshare.com/ndownloader/files/41460054', | |
| # - 'immune':'https://figshare.com/ndownloader/files/41460063', | |
| # | |
| # In[ ]: | |
| ov.utils.download_tosica_gmt() | |
| # ## Initialisation the TOSICA model | |
| # | |
| # We first need to train the TOSICA model on the REFERENCE dataset, where omicverse provides a simple class `pyTOSICA`, and all subsequent operations can be done with `pyTOSICA`. We need to set the parameters for model initialisation. | |
| # | |
| # - `adata`: the reference adata object | |
| # - `gmt_path`: default pre-prepared mask or path to .gmt files. you can use `ov.utils.download_tosica_gmt()` to obtain the genesets | |
| # - `depth`: the depth of transformer model, When it is set to 2, a memory leak may occur | |
| # - `label_name`: the reference key of celltype in `adata.obs` | |
| # - `project_path`: the save path of TOSICA model | |
| # - `batch_size`: indicates the number of cells passed to the programme for training in a single pass | |
| # In[6]: | |
| tosica_obj=ov.single.pyTOSICA(adata=ref_adata, | |
| gmt_path='genesets/GO_bp.gmt', depth=1, | |
| label_name='Celltype', | |
| project_path='hGOBP_demo', | |
| batch_size=8) | |
| # ## Training the TOSICA model | |
| # | |
| # There're 4 arguments to set when training the TOSICA model. | |
| # | |
| # - pre_weights: The path of the pre-trained weights. | |
| # - lr: The learning rate. | |
| # - epochs: The number of epochs. | |
| # - lrf: The learning rate of the last layer. | |
| # In[5]: | |
| tosica_obj.train(epochs=5) | |
| # We can use `.save` to store the `TOSICA` model in `project_path` | |
| # In[6]: | |
| tosica_obj.save() | |
| # The model can be loaded from `project_path` automatically. | |
| # In[7]: | |
| tosica_obj.load() | |
| # ## Update with query | |
| # | |
| # In[8]: | |
| new_adata=tosica_obj.predicted(pre_adata=query_adata) | |
| # ## Visualize the reference and mapping | |
| # | |
| # We first compute the lower dimensional space of query_data, where we use omicverse's preprocessing method as well as the mde method for dimensionality reduction | |
| # | |
| # To visualize the PCA’s embeddings, we use the `pymde` package wrapper in omicverse. This is an alternative to UMAP that is GPU-accelerated. | |
| # In[15]: | |
| ov.pp.scale(query_adata) | |
| ov.pp.pca(query_adata,layer='scaled',n_pcs=50) | |
| sc.pp.neighbors(query_adata, n_neighbors=15, n_pcs=50, | |
| use_rep='scaled|original|X_pca') | |
| query_adata.obsm["X_mde"] = ov.utils.mde(query_adata.obsm["scaled|original|X_pca"]) | |
| query_adata | |
| # Since new_adata and query_adata have the same cells, their low-dimensional spaces are also the same. So we proceed directly to the assignment operation. | |
| # In[16]: | |
| new_adata.obsm=query_adata[new_adata.obs.index].obsm.copy() | |
| new_adata.obsp=query_adata[new_adata.obs.index].obsp.copy() | |
| new_adata | |
| # Since the predicted cell types are not exactly the same as the original cell types, the colours are not exactly the same. For the visualisation effect, we manually set the colour of the predicted cell type with the original cell type. | |
| # In[18]: | |
| import numpy as np | |
| col = np.array([ | |
| "#98DF8A","#E41A1C" ,"#377EB8", "#4DAF4A" ,"#984EA3" ,"#FF7F00" ,"#FFFF33" ,"#A65628" ,"#F781BF" ,"#999999","#1F77B4","#FF7F0E","#279E68","#FF9896" | |
| ]).astype('<U7') | |
| celltype = ("alpha","beta","ductal","acinar","delta","PP","PSC","endothelial","epsilon","mast","macrophage","schwann",'t_cell') | |
| new_adata.obs['Prediction'] = new_adata.obs['Prediction'].astype('category') | |
| new_adata.obs['Prediction'] = new_adata.obs['Prediction'].cat.reorder_categories(list(celltype)) | |
| new_adata.uns['Prediction_colors'] = col[1:] | |
| celltype = ("MHC class II","alpha","beta","ductal","acinar","delta","PP","PSC","endothelial","epsilon","mast") | |
| new_adata.obs['Celltype'] = new_adata.obs['Celltype'].astype('category') | |
| new_adata.obs['Celltype'] = new_adata.obs['Celltype'].cat.reorder_categories(list(celltype)) | |
| new_adata.uns['Celltype_colors'] = col[:11] | |
| # In[24]: | |
| sc.pl.embedding( | |
| new_adata, | |
| basis="X_mde", | |
| color=['Celltype', 'Prediction'], | |
| frameon=False, | |
| #ncols=1, | |
| wspace=0.5, | |
| #palette=ov.utils.pyomic_palette()[11:], | |
| show=False, | |
| ) | |
| # ## Pathway attention | |
| # | |
| # TOSICA has another special feature, which is the ability to computationally use self-attention mechanisms to find pathways associated with cell types. Here we demonstrate the approach of this downstream analysis. | |
| # | |
| # We first need to filter out the predicted types of cells with cell counts less than 5. | |
| # In[30]: | |
| cell_idx=new_adata.obs['Prediction'].value_counts()[new_adata.obs['Prediction'].value_counts()<5].index | |
| new_adata=new_adata[~new_adata.obs['Prediction'].isin(cell_idx)] | |
| # We then used `sc.tl.rank_genes_groups` to calculate the differential pathways with the highest attention across cell types. This differential pathway is derived from the gmt genesets used for the previous calculation. | |
| # In[31]: | |
| sc.tl.rank_genes_groups(new_adata, 'Prediction', method='wilcoxon') | |
| # In[34]: | |
| sc.pl.rank_genes_groups_dotplot(new_adata, | |
| n_genes=3,standard_scale='var',) | |
| # If you want to get the cell-specific pathway, you can use `sc.get.rank_genes_groups_df` to get. | |
| # | |
| # For example, we would like to obtain the pathway with the highest attention for the cell type `PP` | |
| # In[35]: | |
| degs = sc.get.rank_genes_groups_df(new_adata, group='PP', key='rank_genes_groups', | |
| pval_cutoff=0.05) | |
| degs.head() | |
| # In[36]: | |
| sc.pl.embedding( | |
| new_adata, | |
| basis="X_mde", | |
| color=['Prediction','GOBP_REGULATION_OF_MUSCLE_SYSTEM_PROCESS'], | |
| frameon=False, | |
| #ncols=1, | |
| wspace=0.5, | |
| #palette=ov.utils.pyomic_palette()[11:], | |
| show=False, | |
| ) | |
| # If you call omciverse to complete a TOSICA analysis, don't forget to cite the following literature: | |
| # | |
| # ``` | |
| # @article{pmid:36641532, | |
| # journal = {Nature communications}, | |
| # doi = {10.1038/s41467-023-35923-4}, | |
| # issn = {2041-1723}, | |
| # number = {1}, | |
| # pmid = {36641532}, | |
| # pmcid = {PMC9840170}, | |
| # address = {England}, | |
| # title = {Transformer for one stop interpretable cell type annotation}, | |
| # volume = {14}, | |
| # author = {Chen, Jiawei and Xu, Hao and Tao, Wanyu and Chen, Zhaoxiong and Zhao, Yuxuan and Han, Jing-Dong J}, | |
| # note = {[Online; accessed 2023-07-18]}, | |
| # pages = {223}, | |
| # date = {2023-01-14}, | |
| # year = {2023}, | |
| # month = {1}, | |
| # day = {14}, | |
| # } | |
| # | |
| # | |
| # @misc{doi:10.1101/2023.06.06.543913, | |
| # doi = {10.1101/2023.06.06.543913}, | |
| # publisher = {Cold Spring Harbor Laboratory}, | |
| # title = {OmicVerse: A single pipeline for exploring the entire transcriptome universe}, | |
| # author = {Zeng, Zehua and Ma, Yuqing and Hu, Lei and Xiong, Yuanyan and Du, Hongwu}, | |
| # note = {[Online; accessed 2023-07-18]}, | |
| # date = {2023-06-08}, | |
| # year = {2023}, | |
| # month = {6}, | |
| # day = {8}, | |
| # } | |
| # ``` | |
| # In[ ]: | |