haritsahm
commited on
Commit
·
047bb92
1
Parent(s):
68465b3
replace interface with block and remove unused
Browse files- .dockerignore +2 -0
- DESCRIPTION.md +17 -0
- figures/physenet.png +0 -0
- main.py +61 -13
.dockerignore
CHANGED
|
@@ -3,5 +3,7 @@
|
|
| 3 |
!weights/
|
| 4 |
!utils/
|
| 5 |
!examples/
|
|
|
|
| 6 |
!main.py
|
| 7 |
!requirements.txt
|
|
|
|
|
|
| 3 |
!weights/
|
| 4 |
!utils/
|
| 5 |
!examples/
|
| 6 |
+
!figures/
|
| 7 |
!main.py
|
| 8 |
!requirements.txt
|
| 9 |
+
!DESCRIPTION.md
|
DESCRIPTION.md
ADDED
|
@@ -0,0 +1,17 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
## Overview
|
| 2 |
+
|
| 3 |
+
Breast cancer classification using 2-views (bilateral) that compose a mammography exam, owing to the correlations contained in mammography views, which present crucial information for identifying tumors.
|
| 4 |
+
|
| 5 |
+
Reference: https://arxiv.org/abs/2204.05798
|
| 6 |
+
|
| 7 |
+
## Data
|
| 8 |
+
|
| 9 |
+
The model is trained using CBIS [INBReast Dataset](https://pubmed.ncbi.nlm.nih.gov/22078258/)
|
| 10 |
+
|
| 11 |
+
- Target: Lesions (masses, calcifications, asymmetries, and distortions)
|
| 12 |
+
- Task: Segmentation
|
| 13 |
+
- Modality: Grayscale
|
| 14 |
+
|
| 15 |
+
## Demo
|
| 16 |
+
|
| 17 |
+
Please select the example below or upload 2 pairs of mammography exam result.
|
figures/physenet.png
ADDED
|
main.py
CHANGED
|
@@ -21,12 +21,6 @@ BILATERAL_MODEL.eval()
|
|
| 21 |
INPUT_HEIGHT, INPUT_WIDTH = 600, 500
|
| 22 |
|
| 23 |
SUPPORTED_IMG_EXT = ['.png', '.jpg', '.jpeg']
|
| 24 |
-
INPUT_FILES = [
|
| 25 |
-
gr.File(file_count='single', file_types=SUPPORTED_IMG_EXT, label='CC View'),
|
| 26 |
-
gr.File(file_count='single', file_types=SUPPORTED_IMG_EXT, label='MLO View'),
|
| 27 |
-
]
|
| 28 |
-
OUTPUT_GALLERY = gr.Gallery(
|
| 29 |
-
label='Highlighted Area').style(grid=[2], height='auto')
|
| 30 |
EXAMPLE_IMAGES = [
|
| 31 |
['examples/f4b2d377f43ba0bd_left_cc.png',
|
| 32 |
'examples/f4b2d377f43ba0bd_left_mlo.jpg'],
|
|
@@ -186,15 +180,69 @@ def predict_bilateral(cc_file, mlo_file):
|
|
| 186 |
|
| 187 |
def run():
|
| 188 |
"""Run Gradio App."""
|
| 189 |
-
|
| 190 |
-
|
| 191 |
-
|
| 192 |
-
|
| 193 |
-
|
| 194 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 195 |
|
| 196 |
demo.launch(server_name='0.0.0.0', server_port=7860) # nosec B104
|
| 197 |
-
demo.close()
|
| 198 |
|
| 199 |
|
| 200 |
if __name__ == '__main__':
|
|
|
|
| 21 |
INPUT_HEIGHT, INPUT_WIDTH = 600, 500
|
| 22 |
|
| 23 |
SUPPORTED_IMG_EXT = ['.png', '.jpg', '.jpeg']
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 24 |
EXAMPLE_IMAGES = [
|
| 25 |
['examples/f4b2d377f43ba0bd_left_cc.png',
|
| 26 |
'examples/f4b2d377f43ba0bd_left_mlo.jpg'],
|
|
|
|
| 180 |
|
| 181 |
def run():
|
| 182 |
"""Run Gradio App."""
|
| 183 |
+
with open('DESCRIPTION.md', encoding='utf-8') as f:
|
| 184 |
+
description = f.read()
|
| 185 |
+
|
| 186 |
+
with gr.Blocks() as demo:
|
| 187 |
+
with gr.Column():
|
| 188 |
+
gr.Markdown(
|
| 189 |
+
"""
|
| 190 |
+
<h1 style="text-align: center;">Bilateral View Hypercomplex Breast Classification</h1>
|
| 191 |
+
"""
|
| 192 |
+
)
|
| 193 |
+
with gr.Row():
|
| 194 |
+
gr.Markdown(description)
|
| 195 |
+
gr.Markdown(
|
| 196 |
+
"""
|
| 197 |
+
## Model Architecture
|
| 198 |
+
<img src="file/figures/physenet.png" width=auto>
|
| 199 |
+
|
| 200 |
+
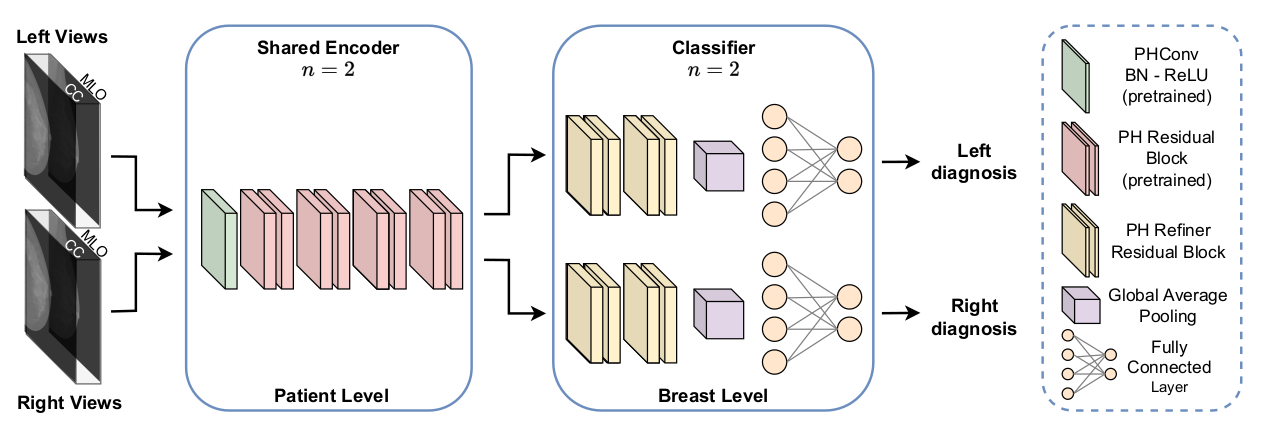
Parameterized Hypercomplex Shared Encoder network (PHYSEnet).
|
| 201 |
+
"""
|
| 202 |
+
)
|
| 203 |
+
with gr.Row():
|
| 204 |
+
with gr.Column():
|
| 205 |
+
cc_file = gr.File(file_count='single',
|
| 206 |
+
file_types=SUPPORTED_IMG_EXT, label='CC View')
|
| 207 |
+
mlo_file = gr.File(file_count='single',
|
| 208 |
+
file_types=SUPPORTED_IMG_EXT, label='MLO View')
|
| 209 |
+
with gr.Row():
|
| 210 |
+
process_btn = gr.Button('Process')
|
| 211 |
+
clear_btn = gr.Button('Clear')
|
| 212 |
+
with gr.Column():
|
| 213 |
+
output_gallery = gr.Gallery(
|
| 214 |
+
label='Highlighted Area').style(grid=[2], height='auto')
|
| 215 |
+
cancer_type = gr.Label(label='Cancer Type')
|
| 216 |
+
gr.Examples(
|
| 217 |
+
examples=EXAMPLE_IMAGES,
|
| 218 |
+
inputs=[cc_file, mlo_file],
|
| 219 |
+
)
|
| 220 |
+
gr.Markdown('Note that this method is sensitive to input image types.\
|
| 221 |
+
Current pipeline expect the values between 0.0-255.0')
|
| 222 |
+
|
| 223 |
+
process_btn.click(
|
| 224 |
+
fn=predict_bilateral,
|
| 225 |
+
inputs=[cc_file, mlo_file],
|
| 226 |
+
outputs=[output_gallery, cancer_type]
|
| 227 |
+
)
|
| 228 |
+
|
| 229 |
+
clear_btn.click(
|
| 230 |
+
lambda _: (
|
| 231 |
+
gr.update(value=None),
|
| 232 |
+
gr.update(value=None),
|
| 233 |
+
gr.update(value=None),
|
| 234 |
+
gr.update(value=None),
|
| 235 |
+
),
|
| 236 |
+
inputs=None,
|
| 237 |
+
outputs=[
|
| 238 |
+
cc_file,
|
| 239 |
+
mlo_file,
|
| 240 |
+
output_gallery,
|
| 241 |
+
cancer_type,
|
| 242 |
+
],
|
| 243 |
+
)
|
| 244 |
|
| 245 |
demo.launch(server_name='0.0.0.0', server_port=7860) # nosec B104
|
|
|
|
| 246 |
|
| 247 |
|
| 248 |
if __name__ == '__main__':
|