Update README Formatting
Browse files- README.md +18 -24
- configs/metadata.json +2 -1
- docs/README.md +18 -24
README.md
CHANGED
|
@@ -5,14 +5,13 @@ tags:
|
|
| 5 |
library_name: monai
|
| 6 |
license: apache-2.0
|
| 7 |
---
|
| 8 |
-
#
|
| 9 |
-
A pre-trained model for classifying nuclei cells as the following types
|
| 10 |
- Other
|
| 11 |
- Inflammatory
|
| 12 |
- Epithelial
|
| 13 |
- Spindle-Shaped
|
| 14 |
|
| 15 |
-
# Model Overview
|
| 16 |
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset.
|
| 17 |
|
| 18 |
## Data
|
|
@@ -23,17 +22,6 @@ unzip -q consep_dataset.zip
|
|
| 23 |
```
|
| 24 |
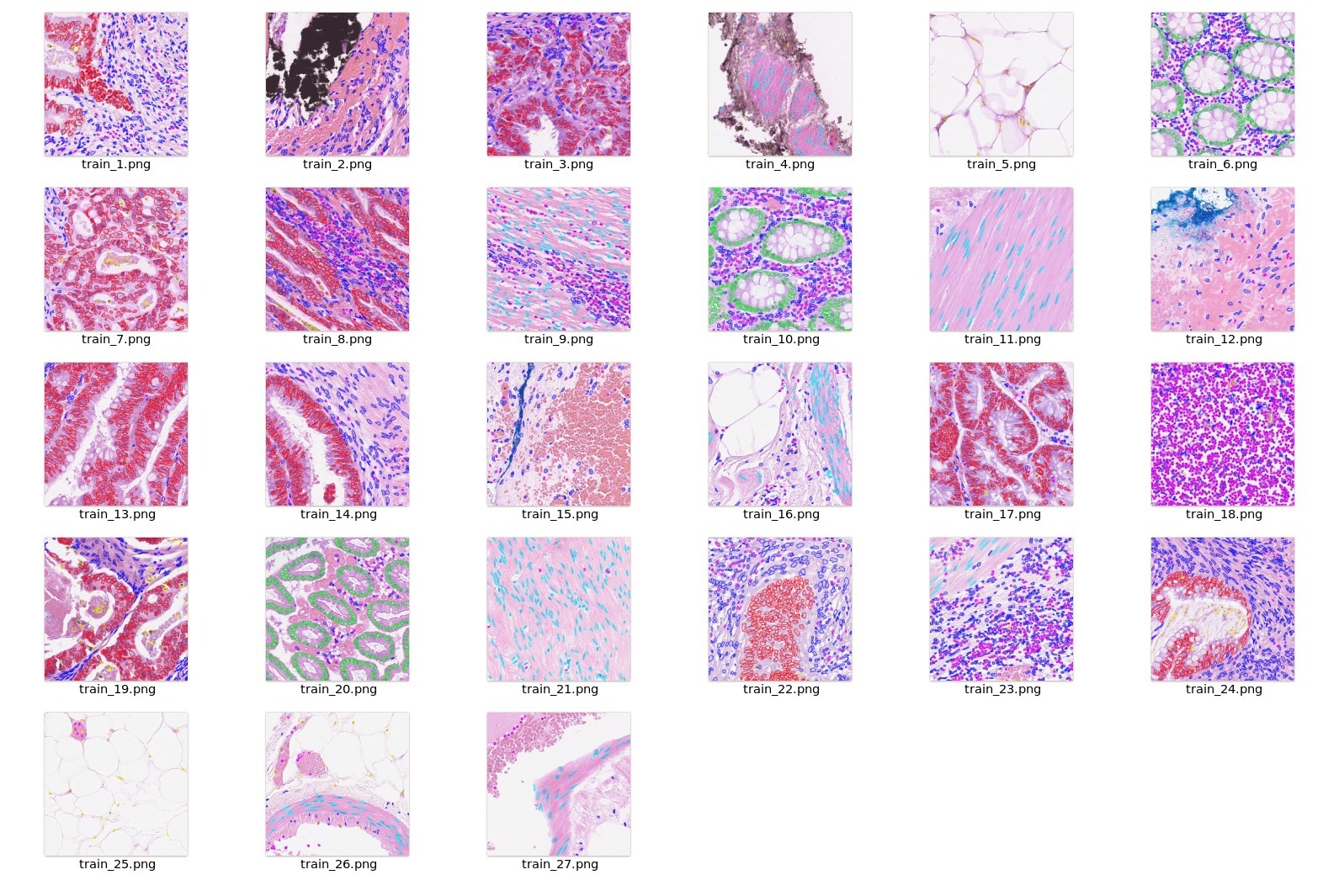<br/>
|
| 25 |
|
| 26 |
-
## Training configuration
|
| 27 |
-
The training was performed with the following:
|
| 28 |
-
|
| 29 |
-
- GPU: at least 12GB of GPU memory
|
| 30 |
-
- Actual Model Input: 4 x 128 x 128
|
| 31 |
-
- AMP: True
|
| 32 |
-
- Optimizer: Adam
|
| 33 |
-
- Learning Rate: 1e-4
|
| 34 |
-
- Loss: torch.nn.CrossEntropyLoss
|
| 35 |
-
|
| 36 |
-
|
| 37 |
### Preprocessing
|
| 38 |
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip),
|
| 39 |
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training.
|
|
@@ -91,13 +79,23 @@ Example `dataset.json` in output folder:
|
|
| 91 |
}
|
| 92 |
```
|
| 93 |
|
|
|
|
|
|
|
| 94 |
|
| 95 |
-
|
| 96 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 97 |
- 3 RGB channels
|
| 98 |
- 1 signal channel (label mask)
|
| 99 |
|
| 100 |
-
|
|
|
|
| 101 |
- 0 = Other
|
| 102 |
- 1 = Inflammatory
|
| 103 |
- 2 = Epithelial
|
|
@@ -132,13 +130,13 @@ Confusion Metrics for <b>Training</b> for individual classes are (at epoch 50):
|
|
| 132 |
|
| 133 |
|
| 134 |
|
| 135 |
-
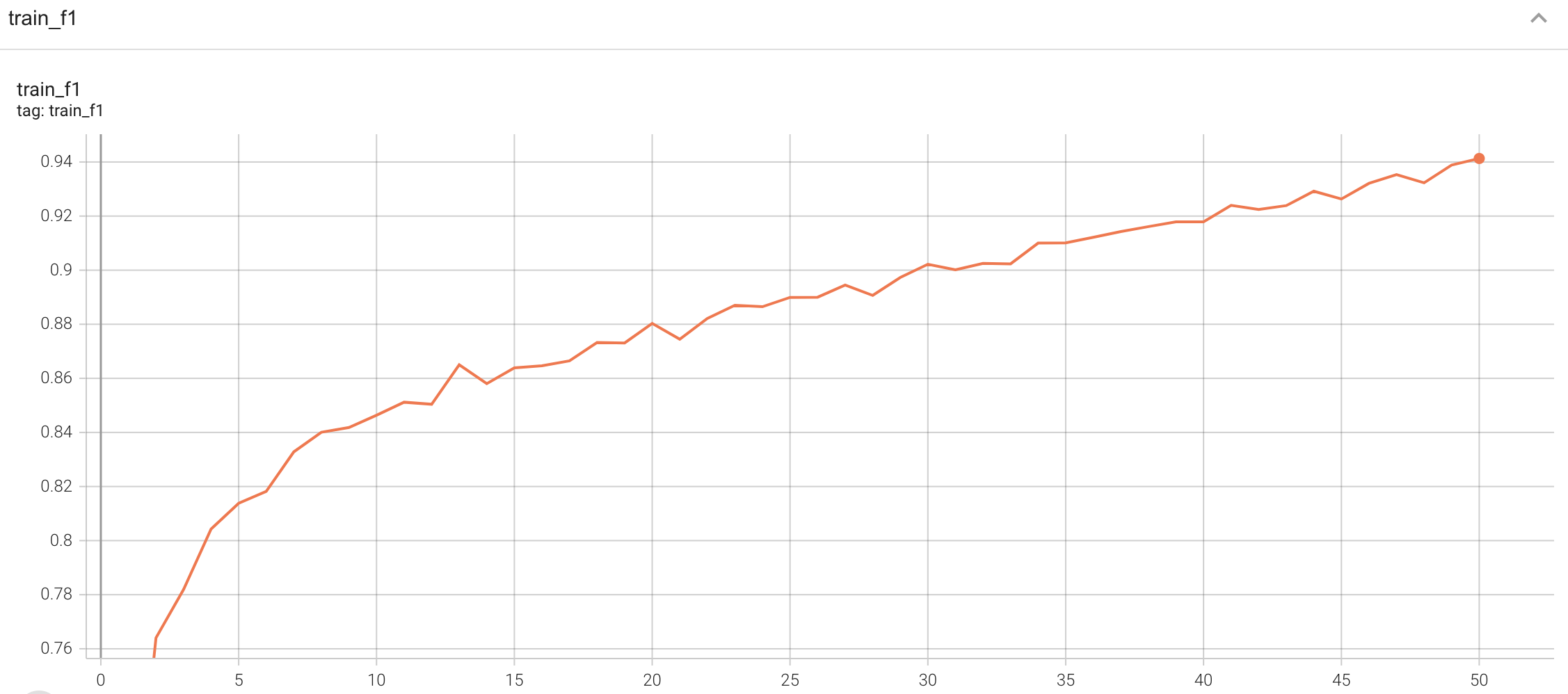
#### Training
|
| 136 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 137 |
|
| 138 |
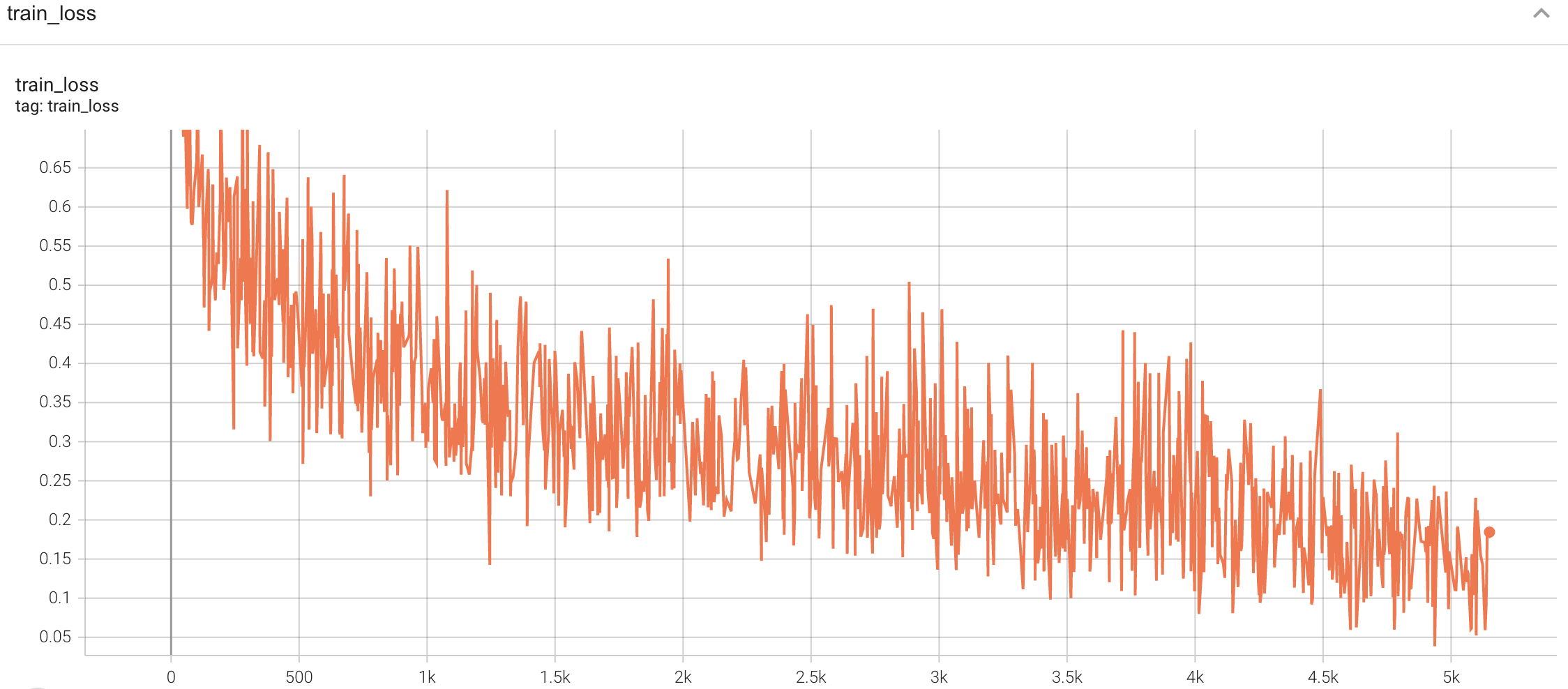 <br>
|
| 139 |
 <br>
|
| 140 |
|
| 141 |
-
#### Validation
|
| 142 |
A graph showing the validation F1-score over 50 epochs.
|
| 143 |
|
| 144 |
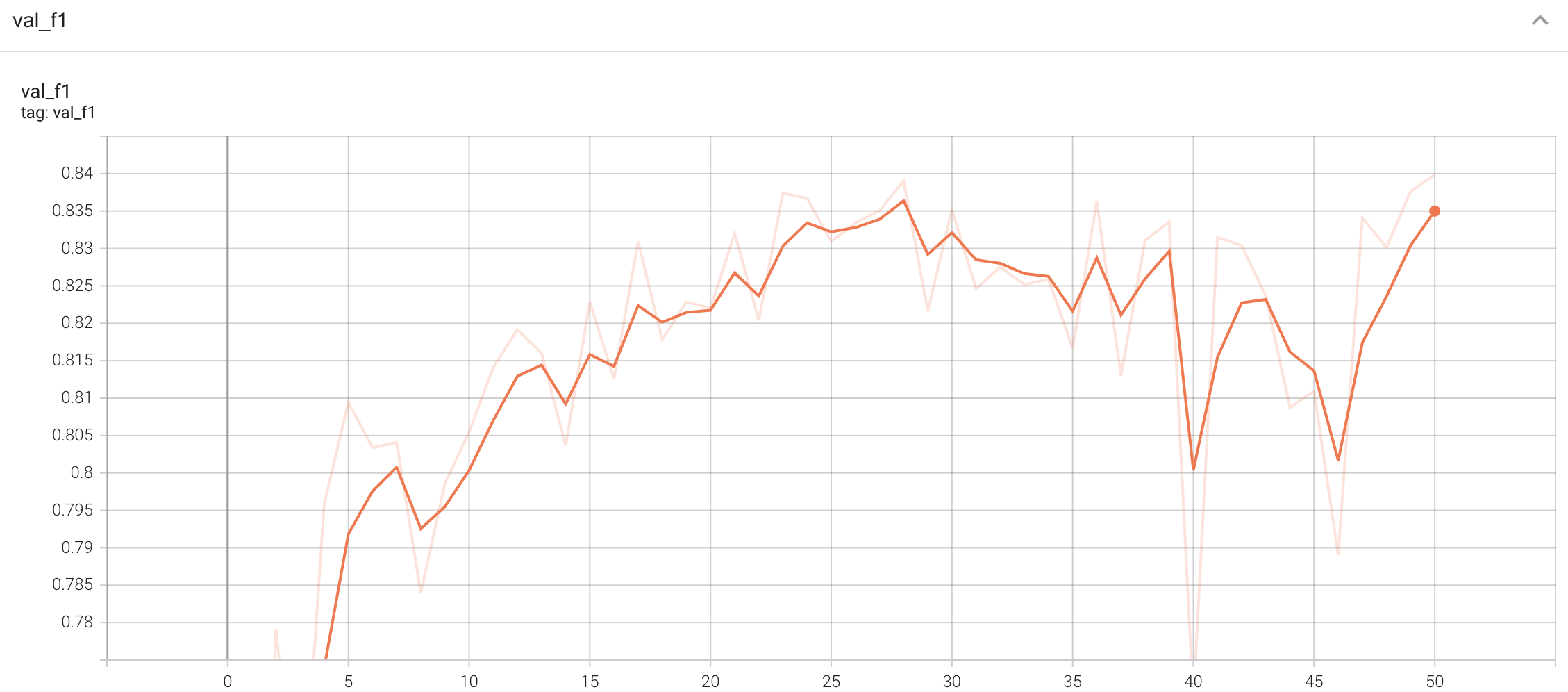 <br>
|
|
@@ -160,8 +158,7 @@ python -m monai.bundle run --config_file configs/train.json
|
|
| 160 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 161 |
```
|
| 162 |
|
| 163 |
-
Please note that the distributed training
|
| 164 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 165 |
|
| 166 |
#### Override the `train` config to execute evaluation with the trained model:
|
| 167 |
|
|
@@ -181,9 +178,6 @@ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config
|
|
| 181 |
python -m monai.bundle run --config_file configs/inference.json
|
| 182 |
```
|
| 183 |
|
| 184 |
-
# Disclaimer
|
| 185 |
-
This is an example, not to be used for diagnostic purposes.
|
| 186 |
-
|
| 187 |
# References
|
| 188 |
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)]
|
| 189 |
|
|
|
|
| 5 |
library_name: monai
|
| 6 |
license: apache-2.0
|
| 7 |
---
|
| 8 |
+
# Model Overview
|
| 9 |
+
A pre-trained model for classifying nuclei cells as the following types
|
| 10 |
- Other
|
| 11 |
- Inflammatory
|
| 12 |
- Epithelial
|
| 13 |
- Spindle-Shaped
|
| 14 |
|
|
|
|
| 15 |
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset.
|
| 16 |
|
| 17 |
## Data
|
|
|
|
| 22 |
```
|
| 23 |
<br/>
|
| 24 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 25 |
### Preprocessing
|
| 26 |
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip),
|
| 27 |
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training.
|
|
|
|
| 79 |
}
|
| 80 |
```
|
| 81 |
|
| 82 |
+
## Training configuration
|
| 83 |
+
The training was performed with the following:
|
| 84 |
|
| 85 |
+
- GPU: at least 12GB of GPU memory
|
| 86 |
+
- Actual Model Input: 4 x 128 x 128
|
| 87 |
+
- AMP: True
|
| 88 |
+
- Optimizer: Adam
|
| 89 |
+
- Learning Rate: 1e-4
|
| 90 |
+
- Loss: torch.nn.CrossEntropyLoss
|
| 91 |
+
|
| 92 |
+
## Input
|
| 93 |
+
4 channels
|
| 94 |
- 3 RGB channels
|
| 95 |
- 1 signal channel (label mask)
|
| 96 |
|
| 97 |
+
## Output
|
| 98 |
+
4 channels
|
| 99 |
- 0 = Other
|
| 100 |
- 1 = Inflammatory
|
| 101 |
- 2 = Epithelial
|
|
|
|
| 130 |
|
| 131 |
|
| 132 |
|
| 133 |
+
#### Training Loss and F1
|
| 134 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 135 |
|
| 136 |
 <br>
|
| 137 |
 <br>
|
| 138 |
|
| 139 |
+
#### Validation F1
|
| 140 |
A graph showing the validation F1-score over 50 epochs.
|
| 141 |
|
| 142 |
 <br>
|
|
|
|
| 158 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 159 |
```
|
| 160 |
|
| 161 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
| 162 |
|
| 163 |
#### Override the `train` config to execute evaluation with the trained model:
|
| 164 |
|
|
|
|
| 178 |
python -m monai.bundle run --config_file configs/inference.json
|
| 179 |
```
|
| 180 |
|
|
|
|
|
|
|
|
|
|
| 181 |
# References
|
| 182 |
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)]
|
| 183 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.0.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.0.8": "enable deterministic training",
|
| 6 |
"0.0.7": "update benchmark on A100",
|
| 7 |
"0.0.6": "adapt to BundleWorkflow interface",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.0.9",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.0.9": "Update README Formatting",
|
| 6 |
"0.0.8": "enable deterministic training",
|
| 7 |
"0.0.7": "update benchmark on A100",
|
| 8 |
"0.0.6": "adapt to BundleWorkflow interface",
|
docs/README.md
CHANGED
|
@@ -1,11 +1,10 @@
|
|
| 1 |
-
#
|
| 2 |
-
A pre-trained model for classifying nuclei cells as the following types
|
| 3 |
- Other
|
| 4 |
- Inflammatory
|
| 5 |
- Epithelial
|
| 6 |
- Spindle-Shaped
|
| 7 |
|
| 8 |
-
# Model Overview
|
| 9 |
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset.
|
| 10 |
|
| 11 |
## Data
|
|
@@ -16,17 +15,6 @@ unzip -q consep_dataset.zip
|
|
| 16 |
```
|
| 17 |
<br/>
|
| 18 |
|
| 19 |
-
## Training configuration
|
| 20 |
-
The training was performed with the following:
|
| 21 |
-
|
| 22 |
-
- GPU: at least 12GB of GPU memory
|
| 23 |
-
- Actual Model Input: 4 x 128 x 128
|
| 24 |
-
- AMP: True
|
| 25 |
-
- Optimizer: Adam
|
| 26 |
-
- Learning Rate: 1e-4
|
| 27 |
-
- Loss: torch.nn.CrossEntropyLoss
|
| 28 |
-
|
| 29 |
-
|
| 30 |
### Preprocessing
|
| 31 |
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip),
|
| 32 |
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training.
|
|
@@ -84,13 +72,23 @@ Example `dataset.json` in output folder:
|
|
| 84 |
}
|
| 85 |
```
|
| 86 |
|
|
|
|
|
|
|
| 87 |
|
| 88 |
-
|
| 89 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 90 |
- 3 RGB channels
|
| 91 |
- 1 signal channel (label mask)
|
| 92 |
|
| 93 |
-
|
|
|
|
| 94 |
- 0 = Other
|
| 95 |
- 1 = Inflammatory
|
| 96 |
- 2 = Epithelial
|
|
@@ -125,13 +123,13 @@ Confusion Metrics for <b>Training</b> for individual classes are (at epoch 50):
|
|
| 125 |
|
| 126 |
|
| 127 |
|
| 128 |
-
#### Training
|
| 129 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 130 |
|
| 131 |
 <br>
|
| 132 |
 <br>
|
| 133 |
|
| 134 |
-
#### Validation
|
| 135 |
A graph showing the validation F1-score over 50 epochs.
|
| 136 |
|
| 137 |
 <br>
|
|
@@ -153,8 +151,7 @@ python -m monai.bundle run --config_file configs/train.json
|
|
| 153 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 154 |
```
|
| 155 |
|
| 156 |
-
Please note that the distributed training
|
| 157 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 158 |
|
| 159 |
#### Override the `train` config to execute evaluation with the trained model:
|
| 160 |
|
|
@@ -174,9 +171,6 @@ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config
|
|
| 174 |
python -m monai.bundle run --config_file configs/inference.json
|
| 175 |
```
|
| 176 |
|
| 177 |
-
# Disclaimer
|
| 178 |
-
This is an example, not to be used for diagnostic purposes.
|
| 179 |
-
|
| 180 |
# References
|
| 181 |
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)]
|
| 182 |
|
|
|
|
| 1 |
+
# Model Overview
|
| 2 |
+
A pre-trained model for classifying nuclei cells as the following types
|
| 3 |
- Other
|
| 4 |
- Inflammatory
|
| 5 |
- Epithelial
|
| 6 |
- Spindle-Shaped
|
| 7 |
|
|
|
|
| 8 |
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset.
|
| 9 |
|
| 10 |
## Data
|
|
|
|
| 15 |
```
|
| 16 |
<br/>
|
| 17 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 18 |
### Preprocessing
|
| 19 |
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip),
|
| 20 |
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training.
|
|
|
|
| 72 |
}
|
| 73 |
```
|
| 74 |
|
| 75 |
+
## Training configuration
|
| 76 |
+
The training was performed with the following:
|
| 77 |
|
| 78 |
+
- GPU: at least 12GB of GPU memory
|
| 79 |
+
- Actual Model Input: 4 x 128 x 128
|
| 80 |
+
- AMP: True
|
| 81 |
+
- Optimizer: Adam
|
| 82 |
+
- Learning Rate: 1e-4
|
| 83 |
+
- Loss: torch.nn.CrossEntropyLoss
|
| 84 |
+
|
| 85 |
+
## Input
|
| 86 |
+
4 channels
|
| 87 |
- 3 RGB channels
|
| 88 |
- 1 signal channel (label mask)
|
| 89 |
|
| 90 |
+
## Output
|
| 91 |
+
4 channels
|
| 92 |
- 0 = Other
|
| 93 |
- 1 = Inflammatory
|
| 94 |
- 2 = Epithelial
|
|
|
|
| 123 |
|
| 124 |
|
| 125 |
|
| 126 |
+
#### Training Loss and F1
|
| 127 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 128 |
|
| 129 |
 <br>
|
| 130 |
 <br>
|
| 131 |
|
| 132 |
+
#### Validation F1
|
| 133 |
A graph showing the validation F1-score over 50 epochs.
|
| 134 |
|
| 135 |
 <br>
|
|
|
|
| 151 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 152 |
```
|
| 153 |
|
| 154 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
| 155 |
|
| 156 |
#### Override the `train` config to execute evaluation with the trained model:
|
| 157 |
|
|
|
|
| 171 |
python -m monai.bundle run --config_file configs/inference.json
|
| 172 |
```
|
| 173 |
|
|
|
|
|
|
|
|
|
|
| 174 |
# References
|
| 175 |
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)]
|
| 176 |
|