Upload 96 files
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +10 -0
- Attention/heatmap/0007_18.nii.gz +3 -0
- Attention/heatmap/0007_18.png +0 -0
- Attention/heatmap/0007_19.nii.gz +3 -0
- Attention/heatmap/0007_19.png +0 -0
- Attention/heatmap/0007_20.nii.gz +3 -0
- Attention/heatmap/0007_20.png +0 -0
- Attention/heatmap/0007_21.nii.gz +3 -0
- Attention/heatmap/0007_21.png +0 -0
- Attention/heatmap/0007_22.nii.gz +3 -0
- Attention/heatmap/0007_22.png +0 -0
- Attention/heatmap/0007_23.nii.gz +3 -0
- Attention/heatmap/0007_23.png +0 -0
- LICENSE +21 -0
- README.md +202 -3
- data/__init__.py +93 -0
- data/aligned_dataset.py +300 -0
- data/base_dataset.py +169 -0
- data/image_folder.py +65 -0
- data/mask_extract.py +346 -0
- data/vertebra_data.json +1468 -0
- datasets/raw/0007/0007.json +50 -0
- datasets/raw/0007/0007.nii.gz +3 -0
- datasets/raw/0007/0007_msk.nii.gz +3 -0
- datasets/straightened/CT/0007_18.nii.gz +3 -0
- datasets/straightened/CT/0007_19.nii.gz +3 -0
- datasets/straightened/CT/0007_20.nii.gz +3 -0
- datasets/straightened/CT/0007_21.nii.gz +3 -0
- datasets/straightened/CT/0007_22.nii.gz +3 -0
- datasets/straightened/CT/0007_23.nii.gz +3 -0
- datasets/straightened/label/0007_18.nii.gz +3 -0
- datasets/straightened/label/0007_19.nii.gz +3 -0
- datasets/straightened/label/0007_20.nii.gz +3 -0
- datasets/straightened/label/0007_21.nii.gz +3 -0
- datasets/straightened/label/0007_22.nii.gz +3 -0
- datasets/straightened/label/0007_23.nii.gz +3 -0
- eval_3d_sagittal_twostage.py +263 -0
- evaluation/RHLV_quantification.py +212 -0
- evaluation/RHLV_quantification_coronal.py +218 -0
- evaluation/SVM_grading.py +96 -0
- evaluation/SVM_grading_2.5d.py +100 -0
- evaluation/generation_eval_coronal.py +175 -0
- evaluation/generation_eval_sagittal.py +165 -0
- images/SHRM_and_HGAM.png +3 -0
- images/attention.png +3 -0
- images/comparison_with_others.png +3 -0
- images/distribution.png +3 -0
- images/mask.png +3 -0
- images/network.png +3 -0
- images/our_method.png +3 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,13 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
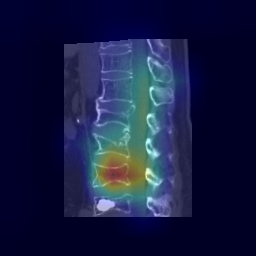
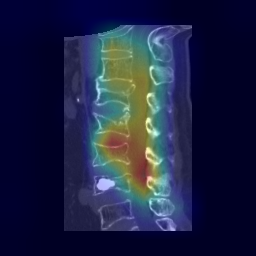
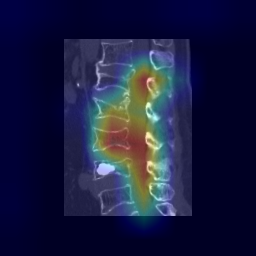
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
images/attention.png filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
images/comparison_with_others.png filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
images/distribution.png filter=lfs diff=lfs merge=lfs -text
|
| 39 |
+
images/mask.png filter=lfs diff=lfs merge=lfs -text
|
| 40 |
+
images/network.png filter=lfs diff=lfs merge=lfs -text
|
| 41 |
+
images/our_method.png filter=lfs diff=lfs merge=lfs -text
|
| 42 |
+
images/rhlv.png filter=lfs diff=lfs merge=lfs -text
|
| 43 |
+
images/SHRM_and_HGAM.png filter=lfs diff=lfs merge=lfs -text
|
| 44 |
+
images/traditional_image_inpaint.png filter=lfs diff=lfs merge=lfs -text
|
| 45 |
+
images/workflow.png filter=lfs diff=lfs merge=lfs -text
|
Attention/heatmap/0007_18.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5c70fe8e6b89d3e406feed30c961de206ebfc17d9f71348e25f6831fdcf228f4
|
| 3 |
+
size 5250286
|
Attention/heatmap/0007_18.png
ADDED

|
Attention/heatmap/0007_19.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e563ebb27cd6259cd98e9c3f99f4ea3c90cfe7a48226b86f69af5cae7215ce11
|
| 3 |
+
size 4993704
|
Attention/heatmap/0007_19.png
ADDED
|
Attention/heatmap/0007_20.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:3693893d1a7ccfa83359291b8784f6c12dca2601c0d081df31c31ef062e758a1
|
| 3 |
+
size 5364097
|
Attention/heatmap/0007_20.png
ADDED

|
Attention/heatmap/0007_21.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:16dcc11eb548ebdbfcc84fccb826a5707b987379b32d10e05056beba6db3fb5a
|
| 3 |
+
size 5114208
|
Attention/heatmap/0007_21.png
ADDED

|
Attention/heatmap/0007_22.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e0a8da6028142d200104a8f747a8acaac39499d36c5a64f982d190e3f9fb194c
|
| 3 |
+
size 4997935
|
Attention/heatmap/0007_22.png
ADDED
|
Attention/heatmap/0007_23.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a4d9b6c372b30477a4875dec40409cde9f0112df8359521942c5f95a7990d0ee
|
| 3 |
+
size 4989910
|
Attention/heatmap/0007_23.png
ADDED
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2025 Qi Zhang
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
CHANGED
|
@@ -1,3 +1,202 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# HealthiVert-GAN: Pseudo-Healthy Vertebral Image Synthesis for Interpretable Compression Fracture Grading
|
| 2 |
+
|
| 3 |
+
[](LICENSE)
|
| 4 |
+
|
| 5 |
+
**HealthiVert-GAN** is a novel framework for synthesizing pseudo-healthy vertebral CT images from fractured vertebrae. By simulating pre-fracture states, it enables interpretable quantification of vertebral compression fractures (VCFs) through **Relative Height Loss of Vertebrae (RHLV)**. The model integrates a two-stage GAN architecture with anatomical consistency modules, achieving state-of-the-art performance on both public and private datasets.
|
| 6 |
+
|
| 7 |
+
---
|
| 8 |
+
|
| 9 |
+
## 🚀 Key Features
|
| 10 |
+
- **Two-Stage Synthesis**: Coarse-to-fine generation with 2.5D sagittal/coronal fusion.
|
| 11 |
+
- **Anatomic Modules**:
|
| 12 |
+
- **Edge-Enhancing Module (EEM)**: Captures precise vertebral morphology.
|
| 13 |
+
- **Self-adaptive Height Restoration Module (SHRM)**: Predicts healthy vertebral height adaptively.
|
| 14 |
+
- **HealthiVert-Guided Attention Module (HGAM)**: Focuses on non-fractured regions via Grad-CAM++.
|
| 15 |
+
- **Iterative Synthesis**: Generates adjacent vertebrae first to minimize fracture interference.
|
| 16 |
+
- **RHLV Quantification**: Measures height loss in anterior/middle/posterior regions for SVM-based Genant grading.
|
| 17 |
+
|
| 18 |
+
---
|
| 19 |
+
|
| 20 |
+
## 🛠️ Architecture
|
| 21 |
+
|
| 22 |
+

|
| 23 |
+
|
| 24 |
+
### Workflow
|
| 25 |
+
1. **Preprocessing**:
|
| 26 |
+
- **Spine Straightening**: Align vertebrae vertically using SCNet segmentation.
|
| 27 |
+
- **De-pedicle**: Remove vertebral arches for body-focused analysis.
|
| 28 |
+
- **Masking**: Replace target vertebra with a fixed-height mask (40mm).
|
| 29 |
+
|
| 30 |
+
2. **Two-Stage Generation**:
|
| 31 |
+
- **Coarse Generator**: Outputs initial CT and segments adjacent vertebrae.
|
| 32 |
+
- **Refinement Generator**: Enhances details with contextual attention and edge loss.
|
| 33 |
+
|
| 34 |
+
3. **Iterative Synthesis**:
|
| 35 |
+
- Step 1: Synthesize adjacent vertebrae.
|
| 36 |
+
- Step 2: Generate target vertebra using Step 1 results.
|
| 37 |
+
|
| 38 |
+
4. **RHLV Calculation**:
|
| 39 |
+
```math
|
| 40 |
+
RHLV = \frac{H_{syn} - H_{ori}}{H_{syn}}
|
| 41 |
+
```
|
| 42 |
+
Segments vertebra into anterior/middle/posterior regions for detailed analysis.
|
| 43 |
+
|
| 44 |
+
**SVM Classification**: Uses RHLV values to classify fractures into mild/moderate/severe.
|
| 45 |
+
|
| 46 |
+
### 🔑 Key Contributions
|
| 47 |
+

|
| 48 |
+
1. **Interpretable Quantification Beyond Black-Box Models**
|
| 49 |
+
Traditional end-to-end fracture classification models suffer from class imbalance and lack interpretability. HealthiVert-GAN addresses these by synthesizing pseudo-healthy vertebrae and quantifying height loss (RHLV) between generated and original vertebrae. This approach achieves superior performance (e.g., **72.3% Macro-F1** on Verse2019) while providing transparent metrics for clinical decisions.
|
| 50 |
+
|
| 51 |
+
2. **Height Loss Distribution Mapping for Surgical Planning**
|
| 52 |
+
HealthiVert-GAN generates cross-sectional height loss heatmaps that visualize compression patterns (wedge/biconcave/crush fractures). Clinicians can use these maps to assess fracture stability and plan interventions (e.g., vertebroplasty) with precision unmatched by single-slice methods.
|
| 53 |
+
|
| 54 |
+
3. **Anatomic Prior Integration**
|
| 55 |
+
Unlike conventional inpainting models, HealthiVert-GAN introduces adjacent vertebrae height variations as prior knowledge. The **Self-adaptive Height Restoration Module (SHRM)** dynamically adjusts generated vertebral heights based on neighboring healthy vertebrae, improving both interpretability and anatomic consistency.
|
| 56 |
+
|
| 57 |
+
|
| 58 |
+
---
|
| 59 |
+
|
| 60 |
+
## 🚀 Quick Start
|
| 61 |
+
|
| 62 |
+
### Installation
|
| 63 |
+
|
| 64 |
+
```bash
|
| 65 |
+
git clone https://github.com/zhibaishouheilab/HealthiVert-GAN.git
|
| 66 |
+
cd HealthiVert-GAN
|
| 67 |
+
pip install -r requirements.txt # PyTorch, NiBabel, SimpleITK, OpenCV
|
| 68 |
+
```
|
| 69 |
+
|
| 70 |
+
### Data Preparation
|
| 71 |
+
|
| 72 |
+
#### Dataset Structure
|
| 73 |
+
Organize data as:
|
| 74 |
+
|
| 75 |
+
```
|
| 76 |
+
/dataset/
|
| 77 |
+
├── raw
|
| 78 |
+
├── 0001/
|
| 79 |
+
│ ├── 0001.nii.gz # Original CT
|
| 80 |
+
│ └── 0001_msk.nii.gz # Vertebrae segmentation
|
| 81 |
+
└── 0002/
|
| 82 |
+
├── 0002.nii.gz
|
| 83 |
+
└── 0002_msk.nii.gz
|
| 84 |
+
```
|
| 85 |
+
**Note**: You have to segment the vertebra firstly. Refer to [CTSpine1K-nnUNet](https://github.com/MIRACLE-Center/CTSpine1K) to obtain how to segment using nnU-Net.
|
| 86 |
+
|
| 87 |
+
#### Preprocessing
|
| 88 |
+
|
| 89 |
+
**Spine Straightening**:
|
| 90 |
+
|
| 91 |
+
```bash
|
| 92 |
+
python straighten/location_json_local.py # Generate vertebral centroids
|
| 93 |
+
python straighten/straighten_mask_3d.py # Output: ./dataset/straightened/
|
| 94 |
+
```
|
| 95 |
+
|
| 96 |
+
**Attention Map Generation**:
|
| 97 |
+
|
| 98 |
+
```bash
|
| 99 |
+
python Attention/grad_CAM_3d_sagittal.py # Output: ./Attention/heatmap/
|
| 100 |
+
```
|
| 101 |
+
|
| 102 |
+
### Training
|
| 103 |
+
|
| 104 |
+
**Configure JSON**:
|
| 105 |
+
|
| 106 |
+
Update `vertebra_data.json` with patient IDs, labels, and paths.
|
| 107 |
+
|
| 108 |
+
**Train Model**:
|
| 109 |
+
|
| 110 |
+
```bash
|
| 111 |
+
python train.py \
|
| 112 |
+
--dataroot ./dataset/straightened \
|
| 113 |
+
--name HealthiVert_experiment \
|
| 114 |
+
--model pix2pix \
|
| 115 |
+
--direction BtoA \
|
| 116 |
+
--batch_size 16 \
|
| 117 |
+
--n_epochs 1000
|
| 118 |
+
```
|
| 119 |
+
|
| 120 |
+
Checkpoints saved in `./checkpoints/HealthiVert_experiment`.
|
| 121 |
+
The pretrained weights will be released later.
|
| 122 |
+
|
| 123 |
+
### Inference
|
| 124 |
+
|
| 125 |
+
**Generate Pseudo-Healthy Vertebrae**:
|
| 126 |
+
|
| 127 |
+
```bash
|
| 128 |
+
python eval_3d_sagittal_twostage.py # define the parameters in the code file
|
| 129 |
+
```
|
| 130 |
+
|
| 131 |
+
Outputs: `./output/CT_fake/` and `./output/label_fake/`.
|
| 132 |
+
|
| 133 |
+
**Fracture Grading**
|
| 134 |
+
|
| 135 |
+
**Calculate RHLV**:
|
| 136 |
+
|
| 137 |
+
```bash
|
| 138 |
+
python evaluation/RHLV_quantification.py
|
| 139 |
+
```
|
| 140 |
+
|
| 141 |
+
**Train SVM Classifier**:
|
| 142 |
+
|
| 143 |
+
```bash
|
| 144 |
+
python evaluation/SVM_grading.py
|
| 145 |
+
```
|
| 146 |
+
|
| 147 |
+
**Evaluate generation results**:
|
| 148 |
+
```bash
|
| 149 |
+
python evaluation/generation_eval_sagittal.py
|
| 150 |
+
```
|
| 151 |
+
---
|
| 152 |
+
|
| 153 |
+
## 📊 Results
|
| 154 |
+
|
| 155 |
+
### Qualitative Comparison
|
| 156 |
+
The generation visulization of different masking strategies.
|
| 157 |
+

|
| 158 |
+
|
| 159 |
+
The visulization heatmap of vertebral height loss distribution in axial view, and the curve of height loss.
|
| 160 |
+

|
| 161 |
+
|
| 162 |
+
### Quantitative Performance (Verse2019 Dataset)
|
| 163 |
+
|
| 164 |
+
| Metric | HealthiVert-GAN | AOT-GAN |3D SupCon-SENet|
|
| 165 |
+
|-------------|-----------------|--------------|------------------|
|
| 166 |
+
| Macro-P | 0.727 | 0.710 | 0.710 |
|
| 167 |
+
| Macro-R | 0.753 | 0.707 | 0.636 |
|
| 168 |
+
| Macro-F1 | 0.723 | 0.692 | 0.667 |
|
| 169 |
+
|
| 170 |
+
Comparison model codes:
|
| 171 |
+
[AOT-GAN](https://github.com/researchmm/AOT-GAN-for-Inpainting)
|
| 172 |
+
[3D SupCon-SENet](https://github.com/wxwxwwxxx/VertebralFractureGrading)
|
| 173 |
+
|
| 174 |
+
---
|
| 175 |
+
|
| 176 |
+
## 📜 Citation
|
| 177 |
+
|
| 178 |
+
```bibtex
|
| 179 |
+
@misc{zhang2025healthivertgannovelframeworkpseudohealthy,
|
| 180 |
+
title={HealthiVert-GAN: A Novel Framework of Pseudo-Healthy Vertebral Image Synthesis for Interpretable Compression Fracture Grading},
|
| 181 |
+
author={Qi Zhang and Shunan Zhang and Ziqi Zhao and Kun Wang and Jun Xu and Jianqi Sun},
|
| 182 |
+
year={2025},
|
| 183 |
+
eprint={2503.05990},
|
| 184 |
+
archivePrefix={arXiv},
|
| 185 |
+
primaryClass={eess.IV},
|
| 186 |
+
url={https://arxiv.org/abs/2503.05990},
|
| 187 |
+
}
|
| 188 |
+
```
|
| 189 |
+
|
| 190 |
+
## 📧 Contact
|
| 191 |
+
If you have any questions about the codes or paper, please let us know via [[email protected]]([email protected]).
|
| 192 |
+
|
| 193 |
+
---
|
| 194 |
+
|
| 195 |
+
## 🙇 Acknowledgment
|
| 196 |
+
- Thank Febian's [nnUnet](https://github.com/MIC-DKFZ/nnUNet).
|
| 197 |
+
- Thank Deng's shared dataset [CTSpine 1K](https://github.com/MIRACLE-Center/CTSpine1K?tab=readme-ov-file) and their pretrained nnUNet's weights.
|
| 198 |
+
- Thank [NeuroML](https://github.com/neuro-ml/straighten) that released the spine straightening algorithm.
|
| 199 |
+
|
| 200 |
+
## 📄 License
|
| 201 |
+
|
| 202 |
+
This project is licensed under the MIT License. See [LICENSE](LICENSE) for details.
|
data/__init__.py
ADDED
|
@@ -0,0 +1,93 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""This package includes all the modules related to data loading and preprocessing
|
| 2 |
+
|
| 3 |
+
To add a custom dataset class called 'dummy', you need to add a file called 'dummy_dataset.py' and define a subclass 'DummyDataset' inherited from BaseDataset.
|
| 4 |
+
You need to implement four functions:
|
| 5 |
+
-- <__init__>: initialize the class, first call BaseDataset.__init__(self, opt).
|
| 6 |
+
-- <__len__>: return the size of dataset.
|
| 7 |
+
-- <__getitem__>: get a data point from data loader.
|
| 8 |
+
-- <modify_commandline_options>: (optionally) add dataset-specific options and set default options.
|
| 9 |
+
|
| 10 |
+
Now you can use the dataset class by specifying flag '--dataset_mode dummy'.
|
| 11 |
+
See our template dataset class 'template_dataset.py' for more details.
|
| 12 |
+
"""
|
| 13 |
+
import importlib
|
| 14 |
+
import torch.utils.data
|
| 15 |
+
from data.base_dataset import BaseDataset
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
def find_dataset_using_name(dataset_name):
|
| 19 |
+
"""Import the module "data/[dataset_name]_dataset.py".
|
| 20 |
+
|
| 21 |
+
In the file, the class called DatasetNameDataset() will
|
| 22 |
+
be instantiated. It has to be a subclass of BaseDataset,
|
| 23 |
+
and it is case-insensitive.
|
| 24 |
+
"""
|
| 25 |
+
dataset_filename = "data." + dataset_name + "_dataset"
|
| 26 |
+
datasetlib = importlib.import_module(dataset_filename)
|
| 27 |
+
|
| 28 |
+
dataset = None
|
| 29 |
+
target_dataset_name = dataset_name.replace('_', '') + 'dataset'
|
| 30 |
+
for name, cls in datasetlib.__dict__.items():
|
| 31 |
+
if name.lower() == target_dataset_name.lower() \
|
| 32 |
+
and issubclass(cls, BaseDataset):
|
| 33 |
+
dataset = cls
|
| 34 |
+
|
| 35 |
+
if dataset is None:
|
| 36 |
+
raise NotImplementedError("In %s.py, there should be a subclass of BaseDataset with class name that matches %s in lowercase." % (dataset_filename, target_dataset_name))
|
| 37 |
+
|
| 38 |
+
return dataset
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
def get_option_setter(dataset_name):
|
| 42 |
+
"""Return the static method <modify_commandline_options> of the dataset class."""
|
| 43 |
+
dataset_class = find_dataset_using_name(dataset_name)
|
| 44 |
+
return dataset_class.modify_commandline_options
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
def create_dataset(opt):
|
| 48 |
+
"""Create a dataset given the option.
|
| 49 |
+
|
| 50 |
+
This function wraps the class CustomDatasetDataLoader.
|
| 51 |
+
This is the main interface between this package and 'train.py'/'test.py'
|
| 52 |
+
|
| 53 |
+
Example:
|
| 54 |
+
>>> from data import create_dataset
|
| 55 |
+
>>> dataset = create_dataset(opt)
|
| 56 |
+
"""
|
| 57 |
+
data_loader = CustomDatasetDataLoader(opt)
|
| 58 |
+
dataset = data_loader.load_data()
|
| 59 |
+
return dataset
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
class CustomDatasetDataLoader():
|
| 63 |
+
"""Wrapper class of Dataset class that performs multi-threaded data loading"""
|
| 64 |
+
|
| 65 |
+
def __init__(self, opt):
|
| 66 |
+
"""Initialize this class
|
| 67 |
+
|
| 68 |
+
Step 1: create a dataset instance given the name [dataset_mode]
|
| 69 |
+
Step 2: create a multi-threaded data loader.
|
| 70 |
+
"""
|
| 71 |
+
self.opt = opt
|
| 72 |
+
dataset_class = find_dataset_using_name(opt.dataset_mode)
|
| 73 |
+
self.dataset = dataset_class(opt)
|
| 74 |
+
print("dataset [%s] was created" % type(self.dataset).__name__)
|
| 75 |
+
self.dataloader = torch.utils.data.DataLoader(
|
| 76 |
+
self.dataset,
|
| 77 |
+
batch_size=opt.batch_size,
|
| 78 |
+
shuffle=not opt.serial_batches,
|
| 79 |
+
num_workers=int(opt.num_threads))
|
| 80 |
+
|
| 81 |
+
def load_data(self):
|
| 82 |
+
return self
|
| 83 |
+
|
| 84 |
+
def __len__(self):
|
| 85 |
+
"""Return the number of data in the dataset"""
|
| 86 |
+
return min(len(self.dataset), self.opt.max_dataset_size)
|
| 87 |
+
|
| 88 |
+
def __iter__(self):
|
| 89 |
+
"""Return a batch of data"""
|
| 90 |
+
for i, data in enumerate(self.dataloader):
|
| 91 |
+
if i * self.opt.batch_size >= self.opt.max_dataset_size:
|
| 92 |
+
break
|
| 93 |
+
yield data
|
data/aligned_dataset.py
ADDED
|
@@ -0,0 +1,300 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#假设读入的数据是nii格式的
|
| 2 |
+
# 用于coronal角度数据的读取
|
| 3 |
+
|
| 4 |
+
import os
|
| 5 |
+
from data.base_dataset import BaseDataset, get_params, get_transform
|
| 6 |
+
from data.image_folder import make_dataset
|
| 7 |
+
from PIL import Image
|
| 8 |
+
import numpy as np
|
| 9 |
+
import torch
|
| 10 |
+
from .mask_extract import process_spine_data, process_spine_data_aug
|
| 11 |
+
import json
|
| 12 |
+
import nibabel as nib
|
| 13 |
+
import random
|
| 14 |
+
import torchvision.transforms as transforms
|
| 15 |
+
from scipy.ndimage import label, find_objects
|
| 16 |
+
|
| 17 |
+
def remove_small_connected_components(input_array, min_size):
|
| 18 |
+
|
| 19 |
+
|
| 20 |
+
# 识别连通域
|
| 21 |
+
structure = np.ones((3, 3), dtype=np.int32) # 定义连通性结构
|
| 22 |
+
labeled, ncomponents = label(input_array, structure)
|
| 23 |
+
|
| 24 |
+
# 遍历所有连通域,如果连通域大小小于阈值,则去除
|
| 25 |
+
for i in range(1, ncomponents + 1):
|
| 26 |
+
if np.sum(labeled == i) < min_size:
|
| 27 |
+
input_array[labeled == i] = 0
|
| 28 |
+
|
| 29 |
+
# 如果输入是张量,则转换回张量
|
| 30 |
+
|
| 31 |
+
return input_array
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
class AlignedDataset(BaseDataset):
|
| 35 |
+
"""A dataset class for paired image dataset.
|
| 36 |
+
|
| 37 |
+
It assumes that the directory '/path/to/data/train' contains image pairs in the form of {A,B}.
|
| 38 |
+
During test time, you need to prepare a directory '/path/to/data/test'.
|
| 39 |
+
"""
|
| 40 |
+
|
| 41 |
+
def __init__(self, opt):
|
| 42 |
+
"""Initialize this dataset class.
|
| 43 |
+
|
| 44 |
+
Parameters:
|
| 45 |
+
opt (Option class) -- stores all the experiment flags; needs to be a subclass of BaseOptions
|
| 46 |
+
"""
|
| 47 |
+
BaseDataset.__init__(self, opt)
|
| 48 |
+
|
| 49 |
+
# 读取json文件来选择训练集、测试集和验证集
|
| 50 |
+
with open('/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/data/vertebra_data.json', 'r') as file:
|
| 51 |
+
vertebra_set = json.load(file)
|
| 52 |
+
self.normal_vert_list = []
|
| 53 |
+
self.abnormal_vert_list = []
|
| 54 |
+
# 初始化存储normal和abnormal vertebrae的字典
|
| 55 |
+
self.normal_vert_dict = {}
|
| 56 |
+
self.abnormal_vert_dict = {}
|
| 57 |
+
|
| 58 |
+
for patient_vert_id in vertebra_set[opt.phase].keys():
|
| 59 |
+
# 分离patient id和vert id
|
| 60 |
+
patient_id, vert_id = patient_vert_id.rsplit('_',1)
|
| 61 |
+
|
| 62 |
+
# 判断该vertebra是normal还是abnormal
|
| 63 |
+
if int(vertebra_set[opt.phase][patient_vert_id]) <= 1:
|
| 64 |
+
self.normal_vert_list.append(patient_vert_id)
|
| 65 |
+
# 如果是normal,添加到normal_vert_dict
|
| 66 |
+
if patient_id not in self.normal_vert_dict:
|
| 67 |
+
self.normal_vert_dict[patient_id] = [vert_id]
|
| 68 |
+
else:
|
| 69 |
+
self.normal_vert_dict[patient_id].append(vert_id)
|
| 70 |
+
else:
|
| 71 |
+
self.abnormal_vert_list.append(patient_vert_id)
|
| 72 |
+
# 如果是abnormal,添加到abnormal_vert_dict
|
| 73 |
+
if patient_id not in self.abnormal_vert_dict:
|
| 74 |
+
self.abnormal_vert_dict[patient_id] = [vert_id]
|
| 75 |
+
else:
|
| 76 |
+
self.abnormal_vert_dict[patient_id].append(vert_id)
|
| 77 |
+
if opt.vert_class=="normal":
|
| 78 |
+
self.vertebra_id = np.array(self.normal_vert_list)
|
| 79 |
+
elif opt.vert_class=="abnormal":
|
| 80 |
+
self.vertebra_id = np.array(self.abnormal_vert_list)
|
| 81 |
+
else:
|
| 82 |
+
print("No vert class is set.")
|
| 83 |
+
self.vertebra_id = None
|
| 84 |
+
|
| 85 |
+
#self.dir_AB = os.path.join(opt.dataroot, opt.phase) # get the image directory
|
| 86 |
+
self.dir_AB = opt.dataroot
|
| 87 |
+
#self.dir_mask = os.path.join(opt.dataroot,'mask',opt.phase)
|
| 88 |
+
#self.AB_paths = sorted(make_dataset(self.dir_AB, opt.max_dataset_size)) # get image paths
|
| 89 |
+
#self.mask_paths = sorted(make_dataset(self.dir_mask, opt.max_dataset_size))
|
| 90 |
+
assert(self.opt.load_size >= self.opt.crop_size) # crop_size should be smaller than the size of loaded image
|
| 91 |
+
self.input_nc = self.opt.output_nc if self.opt.direction == 'BtoA' else self.opt.input_nc
|
| 92 |
+
self.output_nc = self.opt.input_nc if self.opt.direction == 'BtoA' else self.opt.output_nc
|
| 93 |
+
|
| 94 |
+
def numpy_to_pil(self,img_np):
|
| 95 |
+
# 假设 img_np 是一个灰度图像的 NumPy 数组,值域在0到255
|
| 96 |
+
if img_np.dtype != np.uint8:
|
| 97 |
+
raise ValueError("NumPy array should have uint8 data type.")
|
| 98 |
+
# 转换为灰度PIL图像
|
| 99 |
+
img_pil = Image.fromarray(img_np)
|
| 100 |
+
return img_pil
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
|
| 104 |
+
# 按照金字塔概率选择一个slice,毕竟中间的slice包含的信息是最多的,因此尽量选择中间的slice
|
| 105 |
+
# 按照金字塔概率选择一个slice,毕竟中间的slice包含的信息是最多的,因此尽量选择中间的slice
|
| 106 |
+
def get_weighted_random_slice(self,z0, z1):
|
| 107 |
+
# 计算新的范围,限制为原来范围的2/3
|
| 108 |
+
range_length = z1 - z0 + 1
|
| 109 |
+
new_range_length = int(range_length * 4 / 5)
|
| 110 |
+
|
| 111 |
+
# 计算新范围的起始和结束索引
|
| 112 |
+
new_z0 = z0 + (range_length - new_range_length) // 2
|
| 113 |
+
new_z1 = new_z0 + new_range_length - 1
|
| 114 |
+
|
| 115 |
+
# 计算中心索引
|
| 116 |
+
center_index = (new_z0 + new_z1) // 2
|
| 117 |
+
|
| 118 |
+
# 计算每个索引的权重
|
| 119 |
+
weights = [1 - abs(i - center_index) / (new_z1 - new_z0) for i in range(new_z0, new_z1 + 1)]
|
| 120 |
+
|
| 121 |
+
# 归一化权重使得总和为1
|
| 122 |
+
total_weight = sum(weights)
|
| 123 |
+
normalized_weights = [w / total_weight for w in weights]
|
| 124 |
+
|
| 125 |
+
# 根据权重随机选择一个层
|
| 126 |
+
random_index = np.random.choice(range(new_z0, new_z1 + 1), p=normalized_weights)
|
| 127 |
+
index_ratio = abs(random_index-center_index)/range_length*2
|
| 128 |
+
|
| 129 |
+
return random_index,index_ratio
|
| 130 |
+
|
| 131 |
+
def get_valid_slice(self,vert_label, z0, z1,maxheight):
|
| 132 |
+
"""
|
| 133 |
+
尝试随机选取一个非空的slice。
|
| 134 |
+
"""
|
| 135 |
+
max_attempts = 100 # 设定最大尝试次数以避免无限循环
|
| 136 |
+
attempts = 0
|
| 137 |
+
while attempts < max_attempts:
|
| 138 |
+
slice_index,index_ratio = self.get_weighted_random_slice(z0, z1)
|
| 139 |
+
vert_label[:, slice_index, :] = remove_small_connected_components(vert_label[:, slice_index, :],50)
|
| 140 |
+
|
| 141 |
+
if np.sum(vert_label[:, slice_index, :])>50: # 检查切片是否非空
|
| 142 |
+
coords = np.argwhere(vert_label[:, slice_index, :])
|
| 143 |
+
x1, x2 = min(coords[:, 0]), max(coords[:, 0])
|
| 144 |
+
if x2-x1<maxheight:
|
| 145 |
+
return slice_index,index_ratio
|
| 146 |
+
attempts += 1
|
| 147 |
+
raise ValueError("Failed to find a non-empty slice after {} attempts.".format(max_attempts))
|
| 148 |
+
|
| 149 |
+
|
| 150 |
+
def __getitem__(self, index):
|
| 151 |
+
"""Return a data point and its metadata information.
|
| 152 |
+
|
| 153 |
+
Parameters:
|
| 154 |
+
index - - a random integer for data indexing
|
| 155 |
+
|
| 156 |
+
Returns a dictionary that contains A, B, A_paths and B_paths
|
| 157 |
+
A (tensor) - - an image in the input domain
|
| 158 |
+
B (tensor) - - its corresponding image in the target domain
|
| 159 |
+
A_paths (str) - - image paths
|
| 160 |
+
B_paths (str) - - image paths (same as A_paths)
|
| 161 |
+
"""
|
| 162 |
+
# read a image given a random integer index
|
| 163 |
+
CAM_folder = '/home/zhangqi/Project/VertebralFractureGrading/heatmap/straighten_coronal/binaryclass_1'
|
| 164 |
+
CAM_path_0 = os.path.join(CAM_folder, self.vertebra_id[index]+'_0.nii.gz')
|
| 165 |
+
CAM_path_1 = os.path.join(CAM_folder, self.vertebra_id[index]+'_1.nii.gz')
|
| 166 |
+
if not os.path.exists(CAM_path_0):
|
| 167 |
+
CAM_path = CAM_path_1
|
| 168 |
+
else:
|
| 169 |
+
CAM_path = CAM_path_0
|
| 170 |
+
CAM_data = nib.load(CAM_path).get_fdata() * 255
|
| 171 |
+
|
| 172 |
+
|
| 173 |
+
patient_id, vert_id = self.vertebra_id[index].rsplit('_', 1)
|
| 174 |
+
vert_id = int(vert_id)
|
| 175 |
+
normal_vert_list = self.normal_vert_dict[patient_id]
|
| 176 |
+
|
| 177 |
+
|
| 178 |
+
ct_path = os.path.join(self.dir_AB,"CT",self.vertebra_id[index]+'.nii.gz')
|
| 179 |
+
|
| 180 |
+
label_path = os.path.join(self.dir_AB,"label",self.vertebra_id[index]+'.nii.gz')
|
| 181 |
+
|
| 182 |
+
ct_data = nib.load(ct_path).get_fdata()
|
| 183 |
+
label_data = nib.load(label_path).get_fdata()
|
| 184 |
+
vert_label = np.zeros_like(label_data)
|
| 185 |
+
vert_label[label_data==vert_id]=1
|
| 186 |
+
|
| 187 |
+
normal_vert_label = label_data.copy()
|
| 188 |
+
if normal_vert_list:
|
| 189 |
+
for normal_vert in normal_vert_list:
|
| 190 |
+
normal_vert_label[normal_vert_label==int(normal_vert)]=255
|
| 191 |
+
normal_vert_label[normal_vert_label!=255]=0
|
| 192 |
+
else:
|
| 193 |
+
normal_vert_label = np.zeros_like(label_data)
|
| 194 |
+
|
| 195 |
+
loc = np.where(vert_label)
|
| 196 |
+
|
| 197 |
+
# 冠状面选择
|
| 198 |
+
z0 = min(loc[1])
|
| 199 |
+
z1 = max(loc[1])
|
| 200 |
+
maxheight = 40
|
| 201 |
+
|
| 202 |
+
try:
|
| 203 |
+
slice,slice_ratio = self.get_valid_slice(vert_label, z0, z1, maxheight)
|
| 204 |
+
#vert_label[:, :, slice] = remove_small_connected_components(vert_label[:, :, slice],50)
|
| 205 |
+
coords = np.argwhere(vert_label[:, slice, :])
|
| 206 |
+
x1, x2 = min(coords[:, 0]), max(coords[:, 0])
|
| 207 |
+
except ValueError as e:
|
| 208 |
+
print(e)
|
| 209 |
+
width,length = vert_label[:,slice,:].shape
|
| 210 |
+
|
| 211 |
+
height = x2-x1
|
| 212 |
+
mask_x = (x1+x2)//2
|
| 213 |
+
h2 = maxheight
|
| 214 |
+
if height>h2:
|
| 215 |
+
print(slice,ct_path)
|
| 216 |
+
if mask_x<=h2//2:
|
| 217 |
+
min_x = 0
|
| 218 |
+
max_x = min_x + h2
|
| 219 |
+
elif width-mask_x<=h2/2:
|
| 220 |
+
max_x = width
|
| 221 |
+
min_x = max_x -h2
|
| 222 |
+
else:
|
| 223 |
+
min_x = mask_x-h2//2
|
| 224 |
+
max_x = min_x + h2
|
| 225 |
+
|
| 226 |
+
|
| 227 |
+
# 创建256x256的空白数组
|
| 228 |
+
target_A = np.zeros((256, 256))
|
| 229 |
+
target_B = np.zeros((256, 256))
|
| 230 |
+
target_A1 = np.zeros((256, 256))
|
| 231 |
+
target_normal_vert_label = np.zeros((256, 256))
|
| 232 |
+
target_mask = np.zeros((256, 256))
|
| 233 |
+
target_CAM = np.zeros((256, 256))
|
| 234 |
+
|
| 235 |
+
# 定位原切片放置的起始和结束列
|
| 236 |
+
start_col = (256 - 64) // 2
|
| 237 |
+
end_col = start_col + 64
|
| 238 |
+
|
| 239 |
+
# 对于A,直接从ct_data中取切片,然后放置到target_A中
|
| 240 |
+
|
| 241 |
+
target_B[:min_x, start_col:end_col] = ct_data[(x1-min_x):x1, slice, :]
|
| 242 |
+
target_B[max_x:, start_col:end_col] = ct_data[x2:x2+(width-max_x), slice, :]
|
| 243 |
+
|
| 244 |
+
target_A[:, start_col:end_col] = ct_data[:,slice,:]
|
| 245 |
+
|
| 246 |
+
# ���理A1,将label_data中特定ID的位置设为255,其他为0
|
| 247 |
+
A1 = np.zeros_like(label_data[:, slice, :])
|
| 248 |
+
A1[label_data[:, slice, :] == vert_id] = 255
|
| 249 |
+
target_A1[:, start_col:end_col] = A1
|
| 250 |
+
|
| 251 |
+
# 处理normal_vert_label
|
| 252 |
+
target_normal_vert_label[:min_x, start_col:end_col] = normal_vert_label[(x1-min_x):x1, slice, :]
|
| 253 |
+
target_normal_vert_label[max_x:, start_col:end_col] = normal_vert_label[x2:x2+(width-max_x), slice, :]
|
| 254 |
+
|
| 255 |
+
# 处理mask
|
| 256 |
+
target_mask[min_x:max_x, start_col:end_col] = 255
|
| 257 |
+
target_CAM[:min_x, start_col:end_col] = CAM_data[(x1-min_x):x1, slice, :]
|
| 258 |
+
target_CAM[max_x:, start_col:end_col] = CAM_data[x2:x2+(width-max_x), slice, :]
|
| 259 |
+
|
| 260 |
+
target_A = target_A.astype(np.uint8)
|
| 261 |
+
target_B = target_B.astype(np.uint8)
|
| 262 |
+
target_A1 = target_A1.astype(np.uint8)
|
| 263 |
+
target_normal_vert_label = target_normal_vert_label.astype(np.uint8)
|
| 264 |
+
target_mask = target_mask.astype(np.uint8)
|
| 265 |
+
target_CAM = target_CAM.astype(np.uint8)
|
| 266 |
+
|
| 267 |
+
|
| 268 |
+
target_A = self.numpy_to_pil(target_A)
|
| 269 |
+
target_B = self.numpy_to_pil(target_B)
|
| 270 |
+
target_A1 = self.numpy_to_pil(target_A1)
|
| 271 |
+
target_mask = self.numpy_to_pil(target_mask)
|
| 272 |
+
target_normal_vert_label = self.numpy_to_pil(target_normal_vert_label)
|
| 273 |
+
target_CAM = self.numpy_to_pil(target_CAM)
|
| 274 |
+
|
| 275 |
+
# apply the same transform to both A and B
|
| 276 |
+
A_transform =transforms.Compose([
|
| 277 |
+
transforms.Grayscale(1),
|
| 278 |
+
transforms.ToTensor(),
|
| 279 |
+
transforms.Normalize((0.5,), (0.5,))
|
| 280 |
+
])
|
| 281 |
+
|
| 282 |
+
mask_transform = transforms.Compose([
|
| 283 |
+
transforms.ToTensor()
|
| 284 |
+
])
|
| 285 |
+
|
| 286 |
+
target_A = A_transform(target_A)
|
| 287 |
+
target_B = A_transform(target_B)
|
| 288 |
+
target_A1 = mask_transform(target_A1)
|
| 289 |
+
target_mask = mask_transform(target_mask)
|
| 290 |
+
target_normal_vert_label = mask_transform(target_normal_vert_label)
|
| 291 |
+
target_CAM = mask_transform(target_CAM)
|
| 292 |
+
|
| 293 |
+
|
| 294 |
+
|
| 295 |
+
return {'A': target_A, 'A_mask': target_A1, 'mask':target_mask,'B':target_B,'height':height,'x1':x1,'x2':x2,
|
| 296 |
+
'h2':h2,'slice_ratio':slice_ratio,'normal_vert':target_normal_vert_label,'CAM':target_CAM,'A_paths': ct_path, 'B_paths': ct_path}
|
| 297 |
+
|
| 298 |
+
def __len__(self):
|
| 299 |
+
"""Return the total number of images in the dataset."""
|
| 300 |
+
return len(self.vertebra_id)
|
data/base_dataset.py
ADDED
|
@@ -0,0 +1,169 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""This module implements an abstract base class (ABC) 'BaseDataset' for datasets.
|
| 2 |
+
|
| 3 |
+
It also includes common transformation functions (e.g., get_transform, __scale_width), which can be later used in subclasses.
|
| 4 |
+
"""
|
| 5 |
+
import random
|
| 6 |
+
import numpy as np
|
| 7 |
+
import torch.utils.data as data
|
| 8 |
+
from PIL import Image
|
| 9 |
+
import torchvision.transforms as transforms
|
| 10 |
+
from abc import ABC, abstractmethod
|
| 11 |
+
import torch
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
class BaseDataset(data.Dataset, ABC):
|
| 15 |
+
"""This class is an abstract base class (ABC) for datasets.
|
| 16 |
+
|
| 17 |
+
To create a subclass, you need to implement the following four functions:
|
| 18 |
+
-- <__init__>: initialize the class, first call BaseDataset.__init__(self, opt).
|
| 19 |
+
-- <__len__>: return the size of dataset.
|
| 20 |
+
-- <__getitem__>: get a data point.
|
| 21 |
+
-- <modify_commandline_options>: (optionally) add dataset-specific options and set default options.
|
| 22 |
+
"""
|
| 23 |
+
|
| 24 |
+
def __init__(self, opt):
|
| 25 |
+
"""Initialize the class; save the options in the class
|
| 26 |
+
|
| 27 |
+
Parameters:
|
| 28 |
+
opt (Option class)-- stores all the experiment flags; needs to be a subclass of BaseOptions
|
| 29 |
+
"""
|
| 30 |
+
self.opt = opt
|
| 31 |
+
self.root = opt.dataroot
|
| 32 |
+
|
| 33 |
+
@staticmethod
|
| 34 |
+
def modify_commandline_options(parser, is_train):
|
| 35 |
+
"""Add new dataset-specific options, and rewrite default values for existing options.
|
| 36 |
+
|
| 37 |
+
Parameters:
|
| 38 |
+
parser -- original option parser
|
| 39 |
+
is_train (bool) -- whether training phase or test phase. You can use this flag to add training-specific or test-specific options.
|
| 40 |
+
|
| 41 |
+
Returns:
|
| 42 |
+
the modified parser.
|
| 43 |
+
"""
|
| 44 |
+
return parser
|
| 45 |
+
|
| 46 |
+
@abstractmethod
|
| 47 |
+
def __len__(self):
|
| 48 |
+
"""Return the total number of images in the dataset."""
|
| 49 |
+
return 0
|
| 50 |
+
|
| 51 |
+
@abstractmethod
|
| 52 |
+
def __getitem__(self, index):
|
| 53 |
+
"""Return a data point and its metadata information.
|
| 54 |
+
|
| 55 |
+
Parameters:
|
| 56 |
+
index - - a random integer for data indexing
|
| 57 |
+
|
| 58 |
+
Returns:
|
| 59 |
+
a dictionary of data with their names. It ususally contains the data itself and its metadata information.
|
| 60 |
+
"""
|
| 61 |
+
pass
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
def get_params(opt, size):
|
| 65 |
+
w, h = size
|
| 66 |
+
new_h = h
|
| 67 |
+
new_w = w
|
| 68 |
+
if opt.preprocess == 'resize_and_crop':
|
| 69 |
+
new_h = new_w = opt.load_size
|
| 70 |
+
elif opt.preprocess == 'scale_width_and_crop':
|
| 71 |
+
new_w = opt.load_size
|
| 72 |
+
new_h = opt.load_size * h // w
|
| 73 |
+
|
| 74 |
+
x = random.randint(0, np.maximum(0, new_w - opt.crop_size))
|
| 75 |
+
y = random.randint(0, np.maximum(0, new_h - opt.crop_size))
|
| 76 |
+
|
| 77 |
+
flip = random.random() > 0.5
|
| 78 |
+
|
| 79 |
+
return {'crop_pos': (x, y), 'flip': flip}
|
| 80 |
+
|
| 81 |
+
|
| 82 |
+
def get_transform(opt, params=None, grayscale=False, method=transforms.InterpolationMode.BICUBIC, convert=True,normalize=True):
|
| 83 |
+
transform_list = []
|
| 84 |
+
if grayscale:
|
| 85 |
+
transform_list.append(transforms.Grayscale(1))
|
| 86 |
+
if 'resize' in opt.preprocess:
|
| 87 |
+
osize = [opt.load_size, opt.load_size]
|
| 88 |
+
transform_list.append(transforms.Resize(osize, method))
|
| 89 |
+
elif 'scale_width' in opt.preprocess:
|
| 90 |
+
transform_list.append(transforms.Lambda(lambda img: __scale_width(img, opt.load_size, opt.crop_size, method)))
|
| 91 |
+
|
| 92 |
+
if 'crop' in opt.preprocess:
|
| 93 |
+
if params is None:
|
| 94 |
+
transform_list.append(transforms.RandomCrop(opt.crop_size))
|
| 95 |
+
else:
|
| 96 |
+
transform_list.append(transforms.Lambda(lambda img: __crop(img, params['crop_pos'], opt.crop_size)))
|
| 97 |
+
|
| 98 |
+
if opt.preprocess == 'none':
|
| 99 |
+
transform_list.append(transforms.Lambda(lambda img: __make_power_2(img, base=4, method=method)))
|
| 100 |
+
|
| 101 |
+
if not opt.no_flip:
|
| 102 |
+
if params is None:
|
| 103 |
+
transform_list.append(transforms.RandomHorizontalFlip())
|
| 104 |
+
elif params['flip']:
|
| 105 |
+
transform_list.append(transforms.Lambda(lambda img: __flip(img, params['flip'])))
|
| 106 |
+
|
| 107 |
+
if convert:
|
| 108 |
+
transform_list += [transforms.ToTensor()]
|
| 109 |
+
if normalize:
|
| 110 |
+
if grayscale:
|
| 111 |
+
transform_list += [transforms.Normalize((0.5,), (0.5,))]
|
| 112 |
+
else:
|
| 113 |
+
transform_list += [transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))]
|
| 114 |
+
return transforms.Compose(transform_list)
|
| 115 |
+
|
| 116 |
+
|
| 117 |
+
def __transforms2pil_resize(method):
|
| 118 |
+
mapper = {transforms.InterpolationMode.BILINEAR: Image.BILINEAR,
|
| 119 |
+
transforms.InterpolationMode.BICUBIC: Image.BICUBIC,
|
| 120 |
+
transforms.InterpolationMode.NEAREST: Image.NEAREST,
|
| 121 |
+
transforms.InterpolationMode.LANCZOS: Image.LANCZOS,}
|
| 122 |
+
return mapper[method]
|
| 123 |
+
|
| 124 |
+
|
| 125 |
+
def __make_power_2(img, base, method=transforms.InterpolationMode.BICUBIC):
|
| 126 |
+
method = __transforms2pil_resize(method)
|
| 127 |
+
ow, oh = img.size
|
| 128 |
+
h = int(round(oh / base) * base)
|
| 129 |
+
w = int(round(ow / base) * base)
|
| 130 |
+
if h == oh and w == ow:
|
| 131 |
+
return img
|
| 132 |
+
|
| 133 |
+
__print_size_warning(ow, oh, w, h)
|
| 134 |
+
return img.resize((w, h), method)
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
def __scale_width(img, target_size, crop_size, method=transforms.InterpolationMode.BICUBIC):
|
| 138 |
+
method = __transforms2pil_resize(method)
|
| 139 |
+
ow, oh = img.size
|
| 140 |
+
if ow == target_size and oh >= crop_size:
|
| 141 |
+
return img
|
| 142 |
+
w = target_size
|
| 143 |
+
h = int(max(target_size * oh / ow, crop_size))
|
| 144 |
+
return img.resize((w, h), method)
|
| 145 |
+
|
| 146 |
+
|
| 147 |
+
def __crop(img, pos, size):
|
| 148 |
+
ow, oh = img.size
|
| 149 |
+
x1, y1 = pos
|
| 150 |
+
tw = th = size
|
| 151 |
+
if (ow > tw or oh > th):
|
| 152 |
+
return img.crop((x1, y1, x1 + tw, y1 + th))
|
| 153 |
+
return img
|
| 154 |
+
|
| 155 |
+
|
| 156 |
+
def __flip(img, flip):
|
| 157 |
+
if flip:
|
| 158 |
+
return img.transpose(Image.FLIP_LEFT_RIGHT)
|
| 159 |
+
return img
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
def __print_size_warning(ow, oh, w, h):
|
| 163 |
+
"""Print warning information about image size(only print once)"""
|
| 164 |
+
if not hasattr(__print_size_warning, 'has_printed'):
|
| 165 |
+
print("The image size needs to be a multiple of 4. "
|
| 166 |
+
"The loaded image size was (%d, %d), so it was adjusted to "
|
| 167 |
+
"(%d, %d). This adjustment will be done to all images "
|
| 168 |
+
"whose sizes are not multiples of 4" % (ow, oh, w, h))
|
| 169 |
+
__print_size_warning.has_printed = True
|
data/image_folder.py
ADDED
|
@@ -0,0 +1,65 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""A modified image folder class
|
| 2 |
+
|
| 3 |
+
We modify the official PyTorch image folder (https://github.com/pytorch/vision/blob/master/torchvision/datasets/folder.py)
|
| 4 |
+
so that this class can load images from both current directory and its subdirectories.
|
| 5 |
+
"""
|
| 6 |
+
|
| 7 |
+
import torch.utils.data as data
|
| 8 |
+
|
| 9 |
+
from PIL import Image
|
| 10 |
+
import os
|
| 11 |
+
|
| 12 |
+
IMG_EXTENSIONS = [
|
| 13 |
+
'.jpg', '.JPG', '.jpeg', '.JPEG',
|
| 14 |
+
'.png', '.PNG', '.ppm', '.PPM', '.bmp', '.BMP',
|
| 15 |
+
'.tif', '.TIF', '.tiff', '.TIFF',
|
| 16 |
+
]
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
def is_image_file(filename):
|
| 20 |
+
return any(filename.endswith(extension) for extension in IMG_EXTENSIONS)
|
| 21 |
+
|
| 22 |
+
|
| 23 |
+
def make_dataset(dir, max_dataset_size=float("inf")):
|
| 24 |
+
images = []
|
| 25 |
+
assert os.path.isdir(dir), '%s is not a valid directory' % dir
|
| 26 |
+
|
| 27 |
+
for root, _, fnames in sorted(os.walk(dir)):
|
| 28 |
+
for fname in fnames:
|
| 29 |
+
if is_image_file(fname) and "_label" not in fname:
|
| 30 |
+
path = os.path.join(root, fname)
|
| 31 |
+
images.append(path)
|
| 32 |
+
return images[:min(max_dataset_size, len(images))]
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
def default_loader(path):
|
| 36 |
+
return Image.open(path).convert('RGB')
|
| 37 |
+
|
| 38 |
+
|
| 39 |
+
class ImageFolder(data.Dataset):
|
| 40 |
+
|
| 41 |
+
def __init__(self, root, transform=None, return_paths=False,
|
| 42 |
+
loader=default_loader):
|
| 43 |
+
imgs = make_dataset(root)
|
| 44 |
+
if len(imgs) == 0:
|
| 45 |
+
raise(RuntimeError("Found 0 images in: " + root + "\n"
|
| 46 |
+
"Supported image extensions are: " + ",".join(IMG_EXTENSIONS)))
|
| 47 |
+
|
| 48 |
+
self.root = root
|
| 49 |
+
self.imgs = imgs
|
| 50 |
+
self.transform = transform
|
| 51 |
+
self.return_paths = return_paths
|
| 52 |
+
self.loader = loader
|
| 53 |
+
|
| 54 |
+
def __getitem__(self, index):
|
| 55 |
+
path = self.imgs[index]
|
| 56 |
+
img = self.loader(path)
|
| 57 |
+
if self.transform is not None:
|
| 58 |
+
img = self.transform(img)
|
| 59 |
+
if self.return_paths:
|
| 60 |
+
return img, path
|
| 61 |
+
else:
|
| 62 |
+
return img
|
| 63 |
+
|
| 64 |
+
def __len__(self):
|
| 65 |
+
return len(self.imgs)
|
data/mask_extract.py
ADDED
|
@@ -0,0 +1,346 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#coding:utf-8
|
| 2 |
+
import os
|
| 3 |
+
os.environ["KMP_DUPLICATE_LIB_OK"]="TRUE"
|
| 4 |
+
import numpy as np
|
| 5 |
+
import nibabel as nib
|
| 6 |
+
from PIL import Image
|
| 7 |
+
from skimage import morphology
|
| 8 |
+
from skimage.transform import resize
|
| 9 |
+
import cv2
|
| 10 |
+
import os
|
| 11 |
+
import numpy as np
|
| 12 |
+
from skimage import measure
|
| 13 |
+
import skimage
|
| 14 |
+
import numpy.random as npr
|
| 15 |
+
|
| 16 |
+
def get_vertbody(seg0):
|
| 17 |
+
y = []
|
| 18 |
+
count = []
|
| 19 |
+
seg = skimage.morphology.dilation(seg0, skimage.morphology.square(2))
|
| 20 |
+
label, num = measure.label(seg, connectivity=2, background=0, return_num=True)
|
| 21 |
+
out = np.zeros(label.shape)
|
| 22 |
+
loc_list = []
|
| 23 |
+
for i in range(1, num + 1):
|
| 24 |
+
loc = np.where(label == i)
|
| 25 |
+
loc_list.append(loc)
|
| 26 |
+
count.append(loc[0].shape[0])
|
| 27 |
+
y.append(min(list(loc[1])))
|
| 28 |
+
if num == 1:
|
| 29 |
+
print("number=1")
|
| 30 |
+
Num = 0
|
| 31 |
+
countbody = np.sum(label)
|
| 32 |
+
else:
|
| 33 |
+
i = np.argsort(np.array(count))
|
| 34 |
+
if y[i[-1]] < y[i[-2]] or count[i[-2]] < 30:
|
| 35 |
+
|
| 36 |
+
Num = i[-1]
|
| 37 |
+
countbody = count[i[-1]]
|
| 38 |
+
else:
|
| 39 |
+
Num = i[-2]
|
| 40 |
+
countbody = count[i[-2]]
|
| 41 |
+
|
| 42 |
+
out[loc_list[Num]] = 1
|
| 43 |
+
xx = np.max(loc_list[Num][0])
|
| 44 |
+
xi = np.min(loc_list[Num][0])
|
| 45 |
+
yx = np.max(loc_list[Num][1])
|
| 46 |
+
yi = np.min(loc_list[Num][1])
|
| 47 |
+
xm = np.mean(loc_list[Num][0])
|
| 48 |
+
ym = np.mean(loc_list[Num][1])
|
| 49 |
+
out2 = np.zeros((60,60))
|
| 50 |
+
out = out*seg0
|
| 51 |
+
out2[2:3+xx-xi,2:3+yx-yi] = out[xi:xx+1,yi:yx+1]
|
| 52 |
+
return out2,out,np.array([xm,ym])
|
| 53 |
+
|
| 54 |
+
def window(img,win_min,win_max):
|
| 55 |
+
#骨窗窗宽窗位
|
| 56 |
+
imgmax = np.max(img)
|
| 57 |
+
imgmin = np.min(img)
|
| 58 |
+
if imgmax<win_max and imgmin>win_min:
|
| 59 |
+
return img
|
| 60 |
+
for i in range(img.shape[0]):
|
| 61 |
+
img[i] = 255.0 * (img[i] - win_min) / (win_max - win_min)
|
| 62 |
+
min_index = img[i] < 0
|
| 63 |
+
img[i][min_index] = 0
|
| 64 |
+
max_index = img[i] > 255
|
| 65 |
+
img[i][max_index] = 255
|
| 66 |
+
return img
|
| 67 |
+
|
| 68 |
+
# 采取最小旋转矩形框,使用固定scale即不进行扩增
|
| 69 |
+
def process_spine_data(ct_path,label_path,label_id,output_size):
|
| 70 |
+
|
| 71 |
+
# 读取CT数据和标注数据
|
| 72 |
+
#ct_data = nib.load(ct_path).get_fdata()
|
| 73 |
+
#label_data = nib.load(label_path).get_fdata()
|
| 74 |
+
ct_data = np.load(ct_path)
|
| 75 |
+
label_data = np.load(label_path)
|
| 76 |
+
binary_label = label_data.copy()
|
| 77 |
+
binary_label[binary_label!=0]=255
|
| 78 |
+
|
| 79 |
+
|
| 80 |
+
# 进行归一化并*255
|
| 81 |
+
ct_data = window(ct_data, -300, 800)
|
| 82 |
+
|
| 83 |
+
label = int(label_id)
|
| 84 |
+
|
| 85 |
+
|
| 86 |
+
loc = np.where(label_data == label)
|
| 87 |
+
|
| 88 |
+
#if np.isnan(loc[2]):
|
| 89 |
+
# print(ct_path,label)
|
| 90 |
+
|
| 91 |
+
try:
|
| 92 |
+
center_z = int(np.mean(loc[2]))
|
| 93 |
+
except:
|
| 94 |
+
print("发生 ValueError 异常")
|
| 95 |
+
print("loc 的值为:", loc)
|
| 96 |
+
print(ct_path,label)
|
| 97 |
+
_, _, center_z = np.array(np.where(label_data == label)).mean(axis=1).astype(int)
|
| 98 |
+
|
| 99 |
+
|
| 100 |
+
# 对中间层面的椎体去除横突
|
| 101 |
+
label_binary = np.zeros(label_data.shape)
|
| 102 |
+
label_binary[loc] = 1
|
| 103 |
+
y0 = min(loc[1])
|
| 104 |
+
y1 = max(loc[1])
|
| 105 |
+
z0 = min(loc[0])
|
| 106 |
+
z1 = max(loc[0])
|
| 107 |
+
|
| 108 |
+
img2d = label_binary[z0:z1 + 1, y0:y1 + 1, center_z]
|
| 109 |
+
|
| 110 |
+
_, img2d_vertbody, center_point = get_vertbody(img2d)
|
| 111 |
+
|
| 112 |
+
|
| 113 |
+
img2d_vertbody_points = np.where(img2d_vertbody==1)
|
| 114 |
+
img2d_vertbody_aligned=np.zeros_like(label_data[:,:,0], np.uint8)
|
| 115 |
+
# 如果将GT改为生成椎体mask,这样子就不需要纹理灰度信息了
|
| 116 |
+
img2d_vertbody_aligned[img2d_vertbody_points[0]+z0,img2d_vertbody_points[1]+y0]=1
|
| 117 |
+
|
| 118 |
+
# 计算椎体的中心位置
|
| 119 |
+
center_y,center_x = int(np.mean(img2d_vertbody_points[0])+z0),int(np.mean(img2d_vertbody_points[1])+y0)
|
| 120 |
+
|
| 121 |
+
# 截取224x224的矩形框在中心层面
|
| 122 |
+
center_slice = ct_data[:, :, center_z].copy()
|
| 123 |
+
center_label_slice = binary_label[:, :, center_z].copy()
|
| 124 |
+
|
| 125 |
+
# 创建224x224的矩形框
|
| 126 |
+
rect_slice = np.zeros(output_size, dtype=np.uint8)
|
| 127 |
+
rect_label_slice = np.zeros(output_size, dtype=np.uint8)
|
| 128 |
+
|
| 129 |
+
# 计算矩形框的位置
|
| 130 |
+
min_y, max_y = max(0, output_size[0]//2 - center_y), min(output_size[0], output_size[0]//2 + (center_slice.shape[0] - center_y))
|
| 131 |
+
min_x, max_x = max(0, output_size[0]//2 - center_x), min(output_size[0], output_size[0]//2 + (center_slice.shape[1] - center_x))
|
| 132 |
+
|
| 133 |
+
# 将rect_slice放在中间
|
| 134 |
+
rect_slice[min_y:max_y, min_x:max_x] = center_slice[max(center_y - output_size[0]//2, 0):min(center_y + output_size[0]//2, center_slice.shape[0]),
|
| 135 |
+
max(center_x - output_size[0]//2, 0):min(center_x +output_size[0]//2, center_slice.shape[1])]
|
| 136 |
+
|
| 137 |
+
rect_label_slice[min_y:max_y, min_x:max_x] = center_label_slice[max(center_y - output_size[0]//2, 0):min(center_y + output_size[0]//2, center_slice.shape[0]),
|
| 138 |
+
max(center_x - output_size[0]//2, 0):min(center_x + output_size[0]//2, center_slice.shape[1])]
|
| 139 |
+
|
| 140 |
+
# 获取椎体主体的最小旋转矩形
|
| 141 |
+
contours, _ = cv2.findContours(img2d_vertbody_aligned.astype(np.uint8), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
| 142 |
+
rect = cv2.minAreaRect(contours[0])
|
| 143 |
+
|
| 144 |
+
# 将最小旋转矩形的四个顶点转换为整数坐标
|
| 145 |
+
rect_points = np.int0(cv2.boxPoints(rect))
|
| 146 |
+
# 对该最小矩形进行缩放
|
| 147 |
+
# 缩放因子
|
| 148 |
+
scale_factor = 1.2
|
| 149 |
+
center = rect[0]
|
| 150 |
+
scaled_rect_points = ((rect_points - center) * scale_factor) + center
|
| 151 |
+
scaled_rect_points = np.int0(scaled_rect_points)
|
| 152 |
+
|
| 153 |
+
# 创建包围椎体的最小矩形
|
| 154 |
+
bbox_image = np.zeros_like(label_data[:,:,0], np.uint8)
|
| 155 |
+
bbox_cv2 = cv2.cvtColor(bbox_image, cv2.COLOR_GRAY2BGR)
|
| 156 |
+
|
| 157 |
+
cv2.fillPoly(bbox_cv2, [scaled_rect_points], [255,255,255])
|
| 158 |
+
bbox_cv2 = cv2.cvtColor(bbox_cv2, cv2.COLOR_BGR2GRAY)
|
| 159 |
+
|
| 160 |
+
for other_label in range(8, 26): # 假设label范围为1到25
|
| 161 |
+
if other_label != label:
|
| 162 |
+
# 找到其他label的区域
|
| 163 |
+
other_label_locs = np.where(label_data[:,:,center_z] == other_label)
|
| 164 |
+
|
| 165 |
+
# 检查这些区域是否在bbox内,如果在,则将这部分的masked_label设为0
|
| 166 |
+
for y, x in zip(*other_label_locs):
|
| 167 |
+
if bbox_cv2[y, x] == 255: # 如果在bbox内
|
| 168 |
+
bbox_cv2[y, x] = 0 # 将其他label区域设置为0
|
| 169 |
+
|
| 170 |
+
|
| 171 |
+
|
| 172 |
+
masked_image = center_slice.copy()
|
| 173 |
+
masked_image[np.where(bbox_cv2==255)[0],np.where(bbox_cv2==255)[1]] = 0
|
| 174 |
+
masked_label = center_label_slice.copy()
|
| 175 |
+
masked_label[np.where(bbox_cv2==255)[0],np.where(bbox_cv2==255)[1]] = 0
|
| 176 |
+
|
| 177 |
+
masked_slice = np.zeros(output_size, dtype=np.uint8)
|
| 178 |
+
masked_slice[min_y:max_y, min_x:max_x] =masked_image[max(center_y - output_size[0]//2, 0):min(center_y + output_size[0]//2, center_slice.shape[0]),
|
| 179 |
+
max(center_x - output_size[0]//2, 0):min(center_x +output_size[0]//2, center_slice.shape[1])]
|
| 180 |
+
|
| 181 |
+
masked_label_slice = np.zeros(output_size, dtype=np.uint8)
|
| 182 |
+
masked_label_slice[min_y:max_y, min_x:max_x] = masked_label[max(center_y - output_size[0]//2, 0):min(center_y + output_size[0]//2, center_slice.shape[0]),
|
| 183 |
+
max(center_x - output_size[0]//2, 0):min(center_x +output_size[0]//2, center_slice.shape[1])]
|
| 184 |
+
|
| 185 |
+
# 保存mask区域的二值化图像
|
| 186 |
+
mask_binary = np.zeros(output_size, dtype=np.uint8)
|
| 187 |
+
mask_binary[min_y:max_y, min_x:max_x] = bbox_cv2[max(center_y - output_size[0]//2, 0):min(center_y + output_size[0]//2, center_slice.shape[0]),
|
| 188 |
+
max(center_x - output_size[0]//2, 0):min(center_x +output_size[0]//2, center_slice.shape[1])]
|
| 189 |
+
|
| 190 |
+
return rect_slice,rect_label_slice,mask_binary,masked_slice,masked_label_slice
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
def process_spine_data_aug(ct_path,label_path,label_id,output_size):
|
| 194 |
+
|
| 195 |
+
ct_data = np.load(ct_path)
|
| 196 |
+
label_data = np.load(label_path)
|
| 197 |
+
binary_label = label_data.copy()
|
| 198 |
+
binary_label[binary_label!=0]=255
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
# 进行归一化并*255
|
| 202 |
+
ct_data = window(ct_data, -300, 800)
|
| 203 |
+
|
| 204 |
+
label = int(label_id)
|
| 205 |
+
|
| 206 |
+
loc = np.where(label_data == label)
|
| 207 |
+
|
| 208 |
+
try:
|
| 209 |
+
center_z = int(np.mean(loc[2]))
|
| 210 |
+
except:
|
| 211 |
+
print("发生 ValueError 异常")
|
| 212 |
+
print("loc 的值为:", loc)
|
| 213 |
+
print(label_path,label)
|
| 214 |
+
_, _, center_z = np.array(np.where(label_data == label)).mean(axis=1).astype(int)
|
| 215 |
+
|
| 216 |
+
# 对中间层面的椎体去除横突
|
| 217 |
+
label_binary = np.zeros(label_data.shape)
|
| 218 |
+
label_binary[loc] = 1
|
| 219 |
+
y0 = min(loc[1])
|
| 220 |
+
y1 = max(loc[1])
|
| 221 |
+
z0 = min(loc[0])
|
| 222 |
+
z1 = max(loc[0])
|
| 223 |
+
|
| 224 |
+
img2d = label_binary[z0:z1 + 1, y0:y1 + 1, center_z]
|
| 225 |
+
|
| 226 |
+
_, img2d_vertbody, center_point = get_vertbody(img2d)
|
| 227 |
+
|
| 228 |
+
|
| 229 |
+
img2d_vertbody_points = np.where(img2d_vertbody==1)
|
| 230 |
+
img2d_vertbody_aligned=np.zeros_like(label_data[:,:,0], np.uint8)
|
| 231 |
+
# 如果将GT改为生成椎体mask,这样子就不需要纹理灰度信息了
|
| 232 |
+
img2d_vertbody_aligned[img2d_vertbody_points[0]+z0,img2d_vertbody_points[1]+y0]=1
|
| 233 |
+
|
| 234 |
+
# 计算椎体的中心位置
|
| 235 |
+
center_y,center_x = int(np.mean(img2d_vertbody_points[0])+z0),int(np.mean(img2d_vertbody_points[1])+y0)
|
| 236 |
+
|
| 237 |
+
# 截取224x224的矩形框在中心层面
|
| 238 |
+
center_slice = ct_data[:, :, center_z].copy()
|
| 239 |
+
center_label_slice = binary_label[:, :, center_z].copy()
|
| 240 |
+
#center_slice[img2d_vertbody_aligned==1]=255
|
| 241 |
+
|
| 242 |
+
crop_height, crop_width = output_size
|
| 243 |
+
# 计算椎体中心点相对于原始图像边界的最���可移动距离
|
| 244 |
+
max_shift_y = min(center_y, center_slice.shape[0] - center_y, crop_height//2)/2
|
| 245 |
+
max_shift_x = min(center_x, center_slice.shape[1] - center_x, crop_width//2)/2
|
| 246 |
+
|
| 247 |
+
# 随机选择偏移量,保证椎体完全在裁剪图像内
|
| 248 |
+
shift_y = npr.randint(-max_shift_y, max_shift_y + 1)
|
| 249 |
+
shift_x = npr.randint(-max_shift_x, max_shift_x + 1)
|
| 250 |
+
|
| 251 |
+
# 计算随机化后的裁剪起始点
|
| 252 |
+
start_y = center_y + shift_y - crop_height // 2
|
| 253 |
+
start_x = center_x + shift_x - crop_width // 2
|
| 254 |
+
|
| 255 |
+
# 确定裁剪区域在原始图像内的实际位置
|
| 256 |
+
actual_start_y = max(start_y, 0)
|
| 257 |
+
actual_start_x = max(start_x, 0)
|
| 258 |
+
actual_end_y = min(start_y + crop_height, center_slice.shape[0])
|
| 259 |
+
actual_end_x = min(start_x + crop_width, center_slice.shape[1])
|
| 260 |
+
|
| 261 |
+
# 创建224x224的矩形框
|
| 262 |
+
rect_slice = np.zeros(output_size, dtype=np.uint8)
|
| 263 |
+
rect_label_slice = np.zeros(output_size, dtype=np.uint8)
|
| 264 |
+
|
| 265 |
+
# 将原始图像的相应区域复制到裁剪后的图像
|
| 266 |
+
rect_slice[max(-start_y, 0):max(-start_y, 0)+actual_end_y-actual_start_y,
|
| 267 |
+
max(-start_x, 0):max(-start_x, 0)+actual_end_x-actual_start_x] = \
|
| 268 |
+
center_slice[actual_start_y:actual_end_y, actual_start_x:actual_end_x]
|
| 269 |
+
rect_label_slice[max(-start_y, 0):max(-start_y, 0)+actual_end_y-actual_start_y,
|
| 270 |
+
max(-start_x, 0):max(-start_x, 0)+actual_end_x-actual_start_x] = \
|
| 271 |
+
center_label_slice[actual_start_y:actual_end_y, actual_start_x:actual_end_x]
|
| 272 |
+
|
| 273 |
+
# 获取椎体主体的最小旋转矩形
|
| 274 |
+
contours, _ = cv2.findContours(img2d_vertbody_aligned.astype(np.uint8), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
| 275 |
+
rect = cv2.minAreaRect(contours[0])
|
| 276 |
+
contour = contours[0]
|
| 277 |
+
|
| 278 |
+
# 将最小旋转矩形的四个顶点转换为整数坐标
|
| 279 |
+
rect_points = np.int0(cv2.boxPoints(rect))
|
| 280 |
+
|
| 281 |
+
# 对该最小矩形进行缩放
|
| 282 |
+
# 缩放因子
|
| 283 |
+
# 对最小旋转矩形进行1.2-1.4之间的随机缩放
|
| 284 |
+
scale_factor = npr.uniform(1.1, 1.3)
|
| 285 |
+
center = rect[0]
|
| 286 |
+
scaled_rect_points = ((rect_points - center) * scale_factor) + center
|
| 287 |
+
scaled_rect_points = np.int0(scaled_rect_points)
|
| 288 |
+
# 创建包围椎体的最小矩形
|
| 289 |
+
bbox_image = np.zeros_like(label_data[:,:,0], np.uint8)
|
| 290 |
+
bbox_cv2 = cv2.cvtColor(bbox_image, cv2.COLOR_GRAY2BGR)
|
| 291 |
+
|
| 292 |
+
cv2.fillPoly(bbox_cv2, [scaled_rect_points], [255,255,255])
|
| 293 |
+
bbox_cv2 = cv2.cvtColor(bbox_cv2, cv2.COLOR_BGR2GRAY)
|
| 294 |
+
|
| 295 |
+
|
| 296 |
+
# 获取最小外接圆
|
| 297 |
+
#(xc, yc), radius = cv2.minEnclosingCircle(contour)
|
| 298 |
+
#center_circle = (int(xc), int(yc))
|
| 299 |
+
#radius = int(radius*scale_factor)
|
| 300 |
+
|
| 301 |
+
# 绘制最小外接圆到 bbox_cv2 上
|
| 302 |
+
#cv2.circle(bbox_cv2, center_circle, radius, (255), -1) # 用白色填充圆形
|
| 303 |
+
|
| 304 |
+
# 获取最小外接矩形(非旋转)
|
| 305 |
+
#x, y, w, h = cv2.boundingRect(contour)
|
| 306 |
+
# 绘制最小外接矩形到 bbox_cv2 上
|
| 307 |
+
#cv2.rectangle(bbox_cv2, (x, y), (x + w, y + h), (255), -1) # 用白色填充矩形
|
| 308 |
+
|
| 309 |
+
# 应用bbox_cv2后,对label_data进行检查和处理
|
| 310 |
+
# 将bbox内其他label的区域设置为0
|
| 311 |
+
for other_label in range(8, 26): # 假设label范围为1到25
|
| 312 |
+
if other_label != label:
|
| 313 |
+
# 找到其他label的区域
|
| 314 |
+
other_label_locs = np.where(label_data[:,:,center_z] == other_label)
|
| 315 |
+
|
| 316 |
+
# 检查这些区域是否在bbox内,如果在,则将这部分的masked_label设为0
|
| 317 |
+
for y, x in zip(*other_label_locs):
|
| 318 |
+
if bbox_cv2[y, x] == 255: # 如果在bbox内
|
| 319 |
+
bbox_cv2[y, x] = 0 # 将其他label区域设置为0
|
| 320 |
+
|
| 321 |
+
|
| 322 |
+
# 将椎体mask掉
|
| 323 |
+
masked_image = center_slice.copy()
|
| 324 |
+
masked_image[np.where(bbox_cv2==255)[0],np.where(bbox_cv2==255)[1]] = 0
|
| 325 |
+
masked_label = center_label_slice.copy()
|
| 326 |
+
masked_label[np.where(bbox_cv2==255)[0],np.where(bbox_cv2==255)[1]] = 0
|
| 327 |
+
|
| 328 |
+
masked_slice = np.zeros(output_size, dtype=np.uint8)
|
| 329 |
+
masked_slice[max(-start_y, 0):max(-start_y, 0)+actual_end_y-actual_start_y,
|
| 330 |
+
max(-start_x, 0):max(-start_x, 0)+actual_end_x-actual_start_x] =\
|
| 331 |
+
masked_image[actual_start_y:actual_end_y, actual_start_x:actual_end_x]
|
| 332 |
+
|
| 333 |
+
masked_label_slice = np.zeros(output_size, dtype=np.uint8)
|
| 334 |
+
masked_label_slice[max(-start_y, 0):max(-start_y, 0)+actual_end_y-actual_start_y,
|
| 335 |
+
max(-start_x, 0):max(-start_x, 0)+actual_end_x-actual_start_x] = \
|
| 336 |
+
masked_label[actual_start_y:actual_end_y, actual_start_x:actual_end_x]
|
| 337 |
+
|
| 338 |
+
# 保存mask区域的二值化图像
|
| 339 |
+
mask_binary = np.zeros(output_size, dtype=np.uint8)
|
| 340 |
+
mask_binary[max(-start_y, 0):max(-start_y, 0)+actual_end_y-actual_start_y,
|
| 341 |
+
max(-start_x, 0):max(-start_x, 0)+actual_end_x-actual_start_x] = \
|
| 342 |
+
bbox_cv2[actual_start_y:actual_end_y, actual_start_x:actual_end_x]
|
| 343 |
+
|
| 344 |
+
return rect_slice,rect_label_slice,mask_binary,masked_slice,masked_label_slice
|
| 345 |
+
|
| 346 |
+
|
data/vertebra_data.json
ADDED
|
@@ -0,0 +1,1468 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"test": {
|
| 3 |
+
"sub-verse012_17": 0,
|
| 4 |
+
"sub-verse012_18": 0,
|
| 5 |
+
"sub-verse012_19": 0,
|
| 6 |
+
"sub-verse012_20": 3,
|
| 7 |
+
"sub-verse012_21": 0,
|
| 8 |
+
"sub-verse012_22": 1,
|
| 9 |
+
"sub-verse020_8": 0,
|
| 10 |
+
"sub-verse020_9": 0,
|
| 11 |
+
"sub-verse020_10": 0,
|
| 12 |
+
"sub-verse020_11": 0,
|
| 13 |
+
"sub-verse020_12": 1,
|
| 14 |
+
"sub-verse020_13": 2,
|
| 15 |
+
"sub-verse020_14": 0,
|
| 16 |
+
"sub-verse020_15": 3,
|
| 17 |
+
"sub-verse020_16": 3,
|
| 18 |
+
"sub-verse020_17": 0,
|
| 19 |
+
"sub-verse020_18": 3,
|
| 20 |
+
"sub-verse020_19": 0,
|
| 21 |
+
"sub-verse020_20": 3,
|
| 22 |
+
"sub-verse020_21": 2,
|
| 23 |
+
"sub-verse020_22": 2,
|
| 24 |
+
"sub-verse020_23": 0,
|
| 25 |
+
"sub-verse020_24": 0,
|
| 26 |
+
"sub-verse029_8": 0,
|
| 27 |
+
"sub-verse029_9": 0,
|
| 28 |
+
"sub-verse029_10": 0,
|
| 29 |
+
"sub-verse029_11": 0,
|
| 30 |
+
"sub-verse029_12": 0,
|
| 31 |
+
"sub-verse029_13": 0,
|
| 32 |
+
"sub-verse029_14": 0,
|
| 33 |
+
"sub-verse029_15": 0,
|
| 34 |
+
"sub-verse029_16": 0,
|
| 35 |
+
"sub-verse029_17": 0,
|
| 36 |
+
"sub-verse029_18": 0,
|
| 37 |
+
"sub-verse029_19": 0,
|
| 38 |
+
"sub-verse029_20": 0,
|
| 39 |
+
"sub-verse029_21": 0,
|
| 40 |
+
"sub-verse029_22": 0,
|
| 41 |
+
"sub-verse029_23": 0,
|
| 42 |
+
"sub-verse029_24": 0,
|
| 43 |
+
"sub-verse032_19": 0,
|
| 44 |
+
"sub-verse032_20": 0,
|
| 45 |
+
"sub-verse032_21": 0,
|
| 46 |
+
"sub-verse032_22": 0,
|
| 47 |
+
"sub-verse032_23": 0,
|
| 48 |
+
"sub-verse038_18": 0,
|
| 49 |
+
"sub-verse038_19": 0,
|
| 50 |
+
"sub-verse038_20": 0,
|
| 51 |
+
"sub-verse038_21": 0,
|
| 52 |
+
"sub-verse038_22": 0,
|
| 53 |
+
"sub-verse038_23": 2,
|
| 54 |
+
"sub-verse038_24": 0,
|
| 55 |
+
"sub-verse040_16": 0,
|
| 56 |
+
"sub-verse040_17": 0,
|
| 57 |
+
"sub-verse040_18": 0,
|
| 58 |
+
"sub-verse040_19": 1,
|
| 59 |
+
"sub-verse040_20": 3,
|
| 60 |
+
"sub-verse040_21": 1,
|
| 61 |
+
"sub-verse040_22": 1,
|
| 62 |
+
"sub-verse040_23": 0,
|
| 63 |
+
"sub-verse050_8": 0,
|
| 64 |
+
"sub-verse050_9": 0,
|
| 65 |
+
"sub-verse050_10": 0,
|
| 66 |
+
"sub-verse050_11": 0,
|
| 67 |
+
"sub-verse050_12": 0,
|
| 68 |
+
"sub-verse050_13": 0,
|
| 69 |
+
"sub-verse050_14": 0,
|
| 70 |
+
"sub-verse050_15": 0,
|
| 71 |
+
"sub-verse050_16": 0,
|
| 72 |
+
"sub-verse050_17": 0,
|
| 73 |
+
"sub-verse050_18": 0,
|
| 74 |
+
"sub-verse050_19": 0,
|
| 75 |
+
"sub-verse050_20": 0,
|
| 76 |
+
"sub-verse050_21": 0,
|
| 77 |
+
"sub-verse050_22": 0,
|
| 78 |
+
"sub-verse050_23": 0,
|
| 79 |
+
"sub-verse050_24": 0,
|
| 80 |
+
"sub-verse053_20": 0,
|
| 81 |
+
"sub-verse053_21": 2,
|
| 82 |
+
"sub-verse053_22": 0,
|
| 83 |
+
"sub-verse054_20": 0,
|
| 84 |
+
"sub-verse054_21": 2,
|
| 85 |
+
"sub-verse054_22": 1,
|
| 86 |
+
"sub-verse054_23": 0,
|
| 87 |
+
"sub-verse054_24": 2,
|
| 88 |
+
"sub-verse055_14": 0,
|
| 89 |
+
"sub-verse055_15": 0,
|
| 90 |
+
"sub-verse055_16": 0,
|
| 91 |
+
"sub-verse055_17": 0,
|
| 92 |
+
"sub-verse055_18": 0,
|
| 93 |
+
"sub-verse055_19": 0,
|
| 94 |
+
"sub-verse055_20": 0,
|
| 95 |
+
"sub-verse055_21": 0,
|
| 96 |
+
"sub-verse055_22": 0,
|
| 97 |
+
"sub-verse055_23": 0,
|
| 98 |
+
"sub-verse055_24": 0,
|
| 99 |
+
"sub-verse059_14": 0,
|
| 100 |
+
"sub-verse059_15": 0,
|
| 101 |
+
"sub-verse059_16": 2,
|
| 102 |
+
"sub-verse059_17": 0,
|
| 103 |
+
"sub-verse059_18": 0,
|
| 104 |
+
"sub-verse059_19": 2,
|
| 105 |
+
"sub-verse059_20": 1,
|
| 106 |
+
"sub-verse066_20": 1,
|
| 107 |
+
"sub-verse066_21": 0,
|
| 108 |
+
"sub-verse066_22": 0,
|
| 109 |
+
"sub-verse066_23": 0,
|
| 110 |
+
"sub-verse066_24": 0,
|
| 111 |
+
"sub-verse070_16": 0,
|
| 112 |
+
"sub-verse070_17": 0,
|
| 113 |
+
"sub-verse070_18": 1,
|
| 114 |
+
"sub-verse070_19": 1,
|
| 115 |
+
"sub-verse070_20": 3,
|
| 116 |
+
"sub-verse070_21": 0,
|
| 117 |
+
"sub-verse070_22": 0,
|
| 118 |
+
"sub-verse070_23": 0,
|
| 119 |
+
"sub-verse070_24": 0,
|
| 120 |
+
"sub-verse081_18": 1,
|
| 121 |
+
"sub-verse081_19": 1,
|
| 122 |
+
"sub-verse081_20": 3,
|
| 123 |
+
"sub-verse081_21": 1,
|
| 124 |
+
"sub-verse083_20": 1,
|
| 125 |
+
"sub-verse083_21": 0,
|
| 126 |
+
"sub-verse083_22": 0,
|
| 127 |
+
"sub-verse083_23": 1,
|
| 128 |
+
"sub-verse083_24": 0,
|
| 129 |
+
"sub-verse083_25": 0,
|
| 130 |
+
"sub-verse085_20": 1,
|
| 131 |
+
"sub-verse085_21": 1,
|
| 132 |
+
"sub-verse085_22": 0,
|
| 133 |
+
"sub-verse085_23": 0,
|
| 134 |
+
"sub-verse085_24": 0,
|
| 135 |
+
"sub-verse089_18": 0,
|
| 136 |
+
"sub-verse089_19": 0,
|
| 137 |
+
"sub-verse089_20": 0,
|
| 138 |
+
"sub-verse089_21": 0,
|
| 139 |
+
"sub-verse089_22": 0,
|
| 140 |
+
"sub-verse089_23": 1,
|
| 141 |
+
"sub-verse089_24": 0,
|
| 142 |
+
"sub-verse092_8": 0,
|
| 143 |
+
"sub-verse092_9": 0,
|
| 144 |
+
"sub-verse092_10": 0,
|
| 145 |
+
"sub-verse092_11": 0,
|
| 146 |
+
"sub-verse092_12": 0,
|
| 147 |
+
"sub-verse092_13": 0,
|
| 148 |
+
"sub-verse092_14": 0,
|
| 149 |
+
"sub-verse092_15": 0,
|
| 150 |
+
"sub-verse092_16": 0,
|
| 151 |
+
"sub-verse092_17": 2,
|
| 152 |
+
"sub-verse092_18": 3,
|
| 153 |
+
"sub-verse092_19": 1,
|
| 154 |
+
"sub-verse092_20": 0,
|
| 155 |
+
"sub-verse092_21": 0,
|
| 156 |
+
"sub-verse092_22": 0,
|
| 157 |
+
"sub-verse092_23": 0,
|
| 158 |
+
"sub-verse092_24": 0,
|
| 159 |
+
"sub-verse101_8": 0,
|
| 160 |
+
"sub-verse101_9": 0,
|
| 161 |
+
"sub-verse101_10": 0,
|
| 162 |
+
"sub-verse101_11": 0,
|
| 163 |
+
"sub-verse101_12": 0,
|
| 164 |
+
"sub-verse101_13": 0,
|
| 165 |
+
"sub-verse101_14": 0,
|
| 166 |
+
"sub-verse101_15": 0,
|
| 167 |
+
"sub-verse101_16": 0,
|
| 168 |
+
"sub-verse101_17": 0,
|
| 169 |
+
"sub-verse101_18": 0,
|
| 170 |
+
"sub-verse101_19": 0,
|
| 171 |
+
"sub-verse101_20": 0,
|
| 172 |
+
"sub-verse101_21": 0,
|
| 173 |
+
"sub-verse101_22": 0,
|
| 174 |
+
"sub-verse101_23": 0,
|
| 175 |
+
"sub-verse101_24": 0,
|
| 176 |
+
"sub-verse108_8": 0,
|
| 177 |
+
"sub-verse108_9": 0,
|
| 178 |
+
"sub-verse108_10": 0,
|
| 179 |
+
"sub-verse108_11": 0,
|
| 180 |
+
"sub-verse108_12": 0,
|
| 181 |
+
"sub-verse108_13": 0,
|
| 182 |
+
"sub-verse108_14": 0,
|
| 183 |
+
"sub-verse108_15": 0,
|
| 184 |
+
"sub-verse108_16": 0,
|
| 185 |
+
"sub-verse108_17": 0,
|
| 186 |
+
"sub-verse108_18": 0,
|
| 187 |
+
"sub-verse108_19": 0,
|
| 188 |
+
"sub-verse108_20": 0,
|
| 189 |
+
"sub-verse108_21": 0,
|
| 190 |
+
"sub-verse108_22": 0,
|
| 191 |
+
"sub-verse108_23": 0,
|
| 192 |
+
"sub-verse108_24": 0,
|
| 193 |
+
"sub-verse119_8": 0,
|
| 194 |
+
"sub-verse119_9": 0,
|
| 195 |
+
"sub-verse119_10": 1,
|
| 196 |
+
"sub-verse119_11": 1,
|
| 197 |
+
"sub-verse119_12": 0,
|
| 198 |
+
"sub-verse119_13": 0,
|
| 199 |
+
"sub-verse119_14": 1,
|
| 200 |
+
"sub-verse119_15": 2,
|
| 201 |
+
"sub-verse119_16": 0,
|
| 202 |
+
"sub-verse119_17": 0,
|
| 203 |
+
"sub-verse119_18": 0,
|
| 204 |
+
"sub-verse119_19": 2,
|
| 205 |
+
"sub-verse119_20": 0,
|
| 206 |
+
"sub-verse130_15": 0,
|
| 207 |
+
"sub-verse130_16": 0,
|
| 208 |
+
"sub-verse130_17": 2,
|
| 209 |
+
"sub-verse130_18": 1,
|
| 210 |
+
"sub-verse130_19": 3,
|
| 211 |
+
"sub-verse130_20": 0,
|
| 212 |
+
"sub-verse130_21": 2,
|
| 213 |
+
"sub-verse130_22": 0,
|
| 214 |
+
"sub-verse130_23": 0,
|
| 215 |
+
"sub-verse130_24": 0,
|
| 216 |
+
"sub-verse131_17": 0,
|
| 217 |
+
"sub-verse131_18": 0,
|
| 218 |
+
"sub-verse131_19": 1,
|
| 219 |
+
"sub-verse131_20": 2,
|
| 220 |
+
"sub-verse131_21": 0,
|
| 221 |
+
"sub-verse131_22": 0,
|
| 222 |
+
"sub-verse131_23": 0,
|
| 223 |
+
"sub-verse131_24": 0,
|
| 224 |
+
"sub-verse131_25": 0,
|
| 225 |
+
"sub-verse138_16": 0,
|
| 226 |
+
"sub-verse138_17": 0,
|
| 227 |
+
"sub-verse138_18": 0,
|
| 228 |
+
"sub-verse138_19": 0,
|
| 229 |
+
"sub-verse138_20": 0,
|
| 230 |
+
"sub-verse138_21": 0,
|
| 231 |
+
"sub-verse138_22": 0,
|
| 232 |
+
"sub-verse138_23": 0,
|
| 233 |
+
"sub-verse138_24": 0,
|
| 234 |
+
"sub-verse143_20": 0,
|
| 235 |
+
"sub-verse143_21": 2,
|
| 236 |
+
"sub-verse143_22": 0,
|
| 237 |
+
"sub-verse143_23": 0,
|
| 238 |
+
"sub-verse143_24": 0,
|
| 239 |
+
"sub-verse144_16": 0,
|
| 240 |
+
"sub-verse144_17": 0,
|
| 241 |
+
"sub-verse144_21": 3,
|
| 242 |
+
"sub-verse144_22": 2,
|
| 243 |
+
"sub-verse147_20": 1,
|
| 244 |
+
"sub-verse147_21": 0,
|
| 245 |
+
"sub-verse147_22": 2,
|
| 246 |
+
"sub-verse147_23": 0,
|
| 247 |
+
"sub-verse147_24": 0,
|
| 248 |
+
"sub-verse149_17": 0,
|
| 249 |
+
"sub-verse149_18": 0,
|
| 250 |
+
"sub-verse149_19": 3,
|
| 251 |
+
"sub-verse149_20": 2,
|
| 252 |
+
"sub-verse149_21": 1,
|
| 253 |
+
"sub-verse149_22": 2,
|
| 254 |
+
"sub-verse149_23": 2,
|
| 255 |
+
"sub-verse149_24": 0,
|
| 256 |
+
"sub-verse154_16": 0,
|
| 257 |
+
"sub-verse154_17": 0,
|
| 258 |
+
"sub-verse154_18": 0,
|
| 259 |
+
"sub-verse154_19": 0,
|
| 260 |
+
"sub-verse154_20": 0,
|
| 261 |
+
"sub-verse154_21": 0,
|
| 262 |
+
"sub-verse154_22": 0,
|
| 263 |
+
"sub-verse154_23": 0,
|
| 264 |
+
"sub-verse154_24": 0,
|
| 265 |
+
"sub-verse236_8": 0,
|
| 266 |
+
"sub-verse236_9": 0,
|
| 267 |
+
"sub-verse236_10": 0,
|
| 268 |
+
"sub-verse236_11": 1,
|
| 269 |
+
"sub-verse236_12": 1,
|
| 270 |
+
"sub-verse236_13": 0,
|
| 271 |
+
"sub-verse236_14": 0,
|
| 272 |
+
"sub-verse236_15": 0,
|
| 273 |
+
"sub-verse236_16": 0,
|
| 274 |
+
"sub-verse236_17": 0,
|
| 275 |
+
"sub-verse236_18": 0,
|
| 276 |
+
"sub-verse236_19": 0,
|
| 277 |
+
"sub-verse236_20": 0,
|
| 278 |
+
"sub-verse236_21": 0,
|
| 279 |
+
"sub-verse236_22": 0,
|
| 280 |
+
"sub-verse236_23": 0,
|
| 281 |
+
"sub-verse236_24": 0,
|
| 282 |
+
"sub-verse260_9": 1,
|
| 283 |
+
"sub-verse260_10": 1,
|
| 284 |
+
"sub-verse260_11": 0,
|
| 285 |
+
"sub-verse260_12": 0,
|
| 286 |
+
"sub-verse260_13": 0,
|
| 287 |
+
"sub-verse260_14": 0,
|
| 288 |
+
"sub-verse260_15": 0,
|
| 289 |
+
"sub-verse260_16": 1,
|
| 290 |
+
"sub-verse260_17": 0,
|
| 291 |
+
"sub-verse260_18": 1,
|
| 292 |
+
"sub-verse260_19": 0,
|
| 293 |
+
"sub-verse260_20": 0,
|
| 294 |
+
"sub-verse260_21": 1,
|
| 295 |
+
"sub-verse260_22": 0,
|
| 296 |
+
"sub-verse260_23": 0,
|
| 297 |
+
"sub-verse260_24": 0,
|
| 298 |
+
"sub-verse271_8": 0,
|
| 299 |
+
"sub-verse271_9": 0,
|
| 300 |
+
"sub-verse271_10": 0,
|
| 301 |
+
"sub-verse271_11": 0,
|
| 302 |
+
"sub-verse271_12": 0,
|
| 303 |
+
"sub-verse271_13": 0,
|
| 304 |
+
"sub-verse271_14": 0,
|
| 305 |
+
"sub-verse271_15": 0,
|
| 306 |
+
"sub-verse271_16": 0,
|
| 307 |
+
"sub-verse271_17": 0,
|
| 308 |
+
"sub-verse271_18": 0,
|
| 309 |
+
"sub-verse271_19": 0,
|
| 310 |
+
"sub-verse271_20": 2,
|
| 311 |
+
"sub-verse271_21": 0,
|
| 312 |
+
"sub-verse271_22": 0,
|
| 313 |
+
"sub-verse271_23": 0,
|
| 314 |
+
"sub-verse271_24": 0,
|
| 315 |
+
"sub-verse414_series1_8": 0,
|
| 316 |
+
"sub-verse414_series1_9": 0,
|
| 317 |
+
"sub-verse414_series1_10": 0,
|
| 318 |
+
"sub-verse414_series1_11": 0,
|
| 319 |
+
"sub-verse414_series1_12": 0,
|
| 320 |
+
"sub-verse414_series1_13": 0,
|
| 321 |
+
"sub-verse414_series1_14": 1,
|
| 322 |
+
"sub-verse414_series1_15": 1,
|
| 323 |
+
"sub-verse414_series1_16": 0,
|
| 324 |
+
"sub-verse414_series1_17": 0,
|
| 325 |
+
"sub-verse414_series1_18": 0,
|
| 326 |
+
"sub-verse414_series1_19": 0,
|
| 327 |
+
"sub-verse414_series1_20": 0,
|
| 328 |
+
"sub-verse414_series1_21": 0,
|
| 329 |
+
"sub-verse414_series1_22": 0,
|
| 330 |
+
"sub-verse414_series1_23": 0,
|
| 331 |
+
"sub-verse414_series1_24": 0,
|
| 332 |
+
"sub-verse417_series0_8": 0,
|
| 333 |
+
"sub-verse417_series0_9": 0,
|
| 334 |
+
"sub-verse417_series0_10": 0,
|
| 335 |
+
"sub-verse417_series0_11": 0,
|
| 336 |
+
"sub-verse417_series0_12": 0,
|
| 337 |
+
"sub-verse417_series0_13": 0,
|
| 338 |
+
"sub-verse417_series0_14": 0,
|
| 339 |
+
"sub-verse417_series0_15": 0,
|
| 340 |
+
"sub-verse417_series0_16": 0,
|
| 341 |
+
"sub-verse417_series0_17": 0,
|
| 342 |
+
"sub-verse417_series0_18": 0,
|
| 343 |
+
"sub-verse417_series0_19": 0,
|
| 344 |
+
"sub-verse417_series0_20": 0,
|
| 345 |
+
"sub-verse417_series1_18": 0,
|
| 346 |
+
"sub-verse417_series1_19": 0,
|
| 347 |
+
"sub-verse417_series1_20": 0,
|
| 348 |
+
"sub-verse417_series1_21": 0,
|
| 349 |
+
"sub-verse417_series1_22": 0,
|
| 350 |
+
"sub-verse417_series1_23": 0,
|
| 351 |
+
"sub-verse417_series1_24": 0,
|
| 352 |
+
"sub-verse416_series1_8": 1,
|
| 353 |
+
"sub-verse416_series1_9": 1,
|
| 354 |
+
"sub-verse416_series1_10": 1,
|
| 355 |
+
"sub-verse416_series1_11": 0,
|
| 356 |
+
"sub-verse416_series1_12": 0,
|
| 357 |
+
"sub-verse416_series1_13": 0,
|
| 358 |
+
"sub-verse416_series1_14": 0,
|
| 359 |
+
"sub-verse416_series1_15": 0,
|
| 360 |
+
"sub-verse416_series1_16": 0,
|
| 361 |
+
"sub-verse416_series1_17": 0,
|
| 362 |
+
"sub-verse416_series1_18": 0,
|
| 363 |
+
"sub-verse416_series1_19": 0,
|
| 364 |
+
"sub-verse416_series1_20": 0,
|
| 365 |
+
"sub-verse416_series1_21": 0,
|
| 366 |
+
"sub-verse416_series1_22": 0,
|
| 367 |
+
"sub-verse416_series1_23": 0,
|
| 368 |
+
"sub-verse416_series1_24": 0
|
| 369 |
+
},
|
| 370 |
+
"train": {
|
| 371 |
+
"sub-verse004_16": 2,
|
| 372 |
+
"sub-verse004_17": 2,
|
| 373 |
+
"sub-verse004_18": 0,
|
| 374 |
+
"sub-verse004_19": 0,
|
| 375 |
+
"sub-verse004_20": 3,
|
| 376 |
+
"sub-verse004_21": 0,
|
| 377 |
+
"sub-verse004_22": 0,
|
| 378 |
+
"sub-verse005_20": 1,
|
| 379 |
+
"sub-verse005_21": 1,
|
| 380 |
+
"sub-verse005_22": 1,
|
| 381 |
+
"sub-verse005_23": 1,
|
| 382 |
+
"sub-verse005_24": 0,
|
| 383 |
+
"sub-verse006_23": 0,
|
| 384 |
+
"sub-verse006_24": 0,
|
| 385 |
+
"sub-verse007_16": 0,
|
| 386 |
+
"sub-verse007_17": 0,
|
| 387 |
+
"sub-verse007_18": 0,
|
| 388 |
+
"sub-verse007_19": 2,
|
| 389 |
+
"sub-verse007_20": 2,
|
| 390 |
+
"sub-verse007_21": 0,
|
| 391 |
+
"sub-verse007_22": 0,
|
| 392 |
+
"sub-verse008_19": 0,
|
| 393 |
+
"sub-verse008_20": 0,
|
| 394 |
+
"sub-verse008_21": 2,
|
| 395 |
+
"sub-verse008_22": 0,
|
| 396 |
+
"sub-verse008_23": 0,
|
| 397 |
+
"sub-verse008_24": 0,
|
| 398 |
+
"sub-verse009_9": 0,
|
| 399 |
+
"sub-verse009_10": 0,
|
| 400 |
+
"sub-verse009_11": 0,
|
| 401 |
+
"sub-verse009_12": 3,
|
| 402 |
+
"sub-verse009_13": 0,
|
| 403 |
+
"sub-verse009_14": 1,
|
| 404 |
+
"sub-verse009_15": 1,
|
| 405 |
+
"sub-verse009_16": 0,
|
| 406 |
+
"sub-verse009_17": 0,
|
| 407 |
+
"sub-verse009_18": 0,
|
| 408 |
+
"sub-verse009_19": 0,
|
| 409 |
+
"sub-verse009_20": 1,
|
| 410 |
+
"sub-verse009_21": 1,
|
| 411 |
+
"sub-verse009_22": 3,
|
| 412 |
+
"sub-verse009_23": 2,
|
| 413 |
+
"sub-verse009_24": 2,
|
| 414 |
+
"sub-verse014_20": 0,
|
| 415 |
+
"sub-verse014_21": 0,
|
| 416 |
+
"sub-verse014_22": 0,
|
| 417 |
+
"sub-verse014_23": 0,
|
| 418 |
+
"sub-verse014_24": 0,
|
| 419 |
+
"sub-verse015_19": 0,
|
| 420 |
+
"sub-verse015_20": 0,
|
| 421 |
+
"sub-verse015_21": 0,
|
| 422 |
+
"sub-verse031_20": 2,
|
| 423 |
+
"sub-verse031_21": 0,
|
| 424 |
+
"sub-verse031_22": 0,
|
| 425 |
+
"sub-verse031_23": 3,
|
| 426 |
+
"sub-verse031_24": 2,
|
| 427 |
+
"sub-verse033_8": 0,
|
| 428 |
+
"sub-verse033_9": 0,
|
| 429 |
+
"sub-verse033_10": 0,
|
| 430 |
+
"sub-verse033_11": 0,
|
| 431 |
+
"sub-verse033_12": 0,
|
| 432 |
+
"sub-verse033_13": 2,
|
| 433 |
+
"sub-verse033_14": 2,
|
| 434 |
+
"sub-verse033_15": 3,
|
| 435 |
+
"sub-verse033_16": 2,
|
| 436 |
+
"sub-verse033_17": 1,
|
| 437 |
+
"sub-verse033_18": 3,
|
| 438 |
+
"sub-verse033_19": 2,
|
| 439 |
+
"sub-verse033_20": 1,
|
| 440 |
+
"sub-verse033_21": 2,
|
| 441 |
+
"sub-verse033_22": 0,
|
| 442 |
+
"sub-verse033_23": 0,
|
| 443 |
+
"sub-verse033_24": 0,
|
| 444 |
+
"sub-verse034_17": 0,
|
| 445 |
+
"sub-verse034_18": 0,
|
| 446 |
+
"sub-verse034_19": 0,
|
| 447 |
+
"sub-verse034_20": 0,
|
| 448 |
+
"sub-verse034_21": 0,
|
| 449 |
+
"sub-verse034_22": 0,
|
| 450 |
+
"sub-verse034_23": 0,
|
| 451 |
+
"sub-verse034_24": 0,
|
| 452 |
+
"sub-verse036_21": 1,
|
| 453 |
+
"sub-verse043_9": 0,
|
| 454 |
+
"sub-verse043_10": 0,
|
| 455 |
+
"sub-verse043_11": 0,
|
| 456 |
+
"sub-verse043_12": 0,
|
| 457 |
+
"sub-verse043_13": 0,
|
| 458 |
+
"sub-verse043_14": 0,
|
| 459 |
+
"sub-verse043_15": 0,
|
| 460 |
+
"sub-verse043_16": 0,
|
| 461 |
+
"sub-verse043_17": 0,
|
| 462 |
+
"sub-verse043_18": 0,
|
| 463 |
+
"sub-verse043_19": 2,
|
| 464 |
+
"sub-verse043_20": 0,
|
| 465 |
+
"sub-verse043_21": 0,
|
| 466 |
+
"sub-verse043_22": 0,
|
| 467 |
+
"sub-verse043_23": 0,
|
| 468 |
+
"sub-verse043_24": 0,
|
| 469 |
+
"sub-verse046_20": 0,
|
| 470 |
+
"sub-verse046_21": 0,
|
| 471 |
+
"sub-verse046_22": 1,
|
| 472 |
+
"sub-verse046_23": 1,
|
| 473 |
+
"sub-verse046_24": 0,
|
| 474 |
+
"sub-verse048_21": 0,
|
| 475 |
+
"sub-verse048_22": 0,
|
| 476 |
+
"sub-verse048_23": 0,
|
| 477 |
+
"sub-verse048_24": 0,
|
| 478 |
+
"sub-verse051_17": 0,
|
| 479 |
+
"sub-verse051_18": 0,
|
| 480 |
+
"sub-verse051_19": 0,
|
| 481 |
+
"sub-verse051_20": 0,
|
| 482 |
+
"sub-verse051_21": 0,
|
| 483 |
+
"sub-verse051_22": 0,
|
| 484 |
+
"sub-verse051_23": 0,
|
| 485 |
+
"sub-verse051_24": 0,
|
| 486 |
+
"sub-verse056_18": 0,
|
| 487 |
+
"sub-verse056_19": 2,
|
| 488 |
+
"sub-verse056_23": 2,
|
| 489 |
+
"sub-verse060_19": 0,
|
| 490 |
+
"sub-verse060_20": 2,
|
| 491 |
+
"sub-verse060_21": 1,
|
| 492 |
+
"sub-verse060_22": 2,
|
| 493 |
+
"sub-verse060_23": 1,
|
| 494 |
+
"sub-verse060_24": 3,
|
| 495 |
+
"sub-verse061_17": 0,
|
| 496 |
+
"sub-verse061_18": 0,
|
| 497 |
+
"sub-verse061_19": 0,
|
| 498 |
+
"sub-verse061_20": 2,
|
| 499 |
+
"sub-verse061_21": 2,
|
| 500 |
+
"sub-verse061_22": 0,
|
| 501 |
+
"sub-verse061_23": 0,
|
| 502 |
+
"sub-verse061_24": 0,
|
| 503 |
+
"sub-verse063_20": 0,
|
| 504 |
+
"sub-verse064_9": 0,
|
| 505 |
+
"sub-verse064_10": 0,
|
| 506 |
+
"sub-verse064_11": 0,
|
| 507 |
+
"sub-verse064_12": 0,
|
| 508 |
+
"sub-verse064_13": 1,
|
| 509 |
+
"sub-verse064_14": 0,
|
| 510 |
+
"sub-verse064_15": 0,
|
| 511 |
+
"sub-verse064_16": 0,
|
| 512 |
+
"sub-verse064_17": 0,
|
| 513 |
+
"sub-verse064_18": 0,
|
| 514 |
+
"sub-verse064_19": 0,
|
| 515 |
+
"sub-verse064_20": 0,
|
| 516 |
+
"sub-verse064_21": 2,
|
| 517 |
+
"sub-verse064_22": 0,
|
| 518 |
+
"sub-verse064_23": 0,
|
| 519 |
+
"sub-verse064_24": 0,
|
| 520 |
+
"sub-verse065_21": 0,
|
| 521 |
+
"sub-verse065_22": 0,
|
| 522 |
+
"sub-verse065_23": 0,
|
| 523 |
+
"sub-verse065_24": 0,
|
| 524 |
+
"sub-verse068_16": 0,
|
| 525 |
+
"sub-verse068_17": 0,
|
| 526 |
+
"sub-verse068_18": 0,
|
| 527 |
+
"sub-verse068_19": 0,
|
| 528 |
+
"sub-verse068_20": 3,
|
| 529 |
+
"sub-verse068_21": 3,
|
| 530 |
+
"sub-verse068_22": 0,
|
| 531 |
+
"sub-verse068_23": 0,
|
| 532 |
+
"sub-verse068_24": 0,
|
| 533 |
+
"sub-verse072_19": 0,
|
| 534 |
+
"sub-verse072_20": 0,
|
| 535 |
+
"sub-verse072_21": 1,
|
| 536 |
+
"sub-verse072_22": 0,
|
| 537 |
+
"sub-verse072_23": 0,
|
| 538 |
+
"sub-verse072_24": 0,
|
| 539 |
+
"sub-verse074_8": 0,
|
| 540 |
+
"sub-verse074_9": 0,
|
| 541 |
+
"sub-verse074_10": 0,
|
| 542 |
+
"sub-verse074_11": 0,
|
| 543 |
+
"sub-verse074_12": 0,
|
| 544 |
+
"sub-verse074_13": 0,
|
| 545 |
+
"sub-verse074_14": 0,
|
| 546 |
+
"sub-verse074_15": 0,
|
| 547 |
+
"sub-verse074_16": 0,
|
| 548 |
+
"sub-verse074_17": 0,
|
| 549 |
+
"sub-verse074_18": 0,
|
| 550 |
+
"sub-verse074_19": 0,
|
| 551 |
+
"sub-verse074_20": 0,
|
| 552 |
+
"sub-verse074_21": 0,
|
| 553 |
+
"sub-verse074_22": 0,
|
| 554 |
+
"sub-verse074_23": 0,
|
| 555 |
+
"sub-verse074_24": 0,
|
| 556 |
+
"sub-verse075_16": 0,
|
| 557 |
+
"sub-verse075_17": 0,
|
| 558 |
+
"sub-verse075_18": 0,
|
| 559 |
+
"sub-verse075_19": 0,
|
| 560 |
+
"sub-verse075_20": 0,
|
| 561 |
+
"sub-verse075_21": 0,
|
| 562 |
+
"sub-verse075_22": 0,
|
| 563 |
+
"sub-verse076_20": 0,
|
| 564 |
+
"sub-verse076_21": 0,
|
| 565 |
+
"sub-verse076_22": 1,
|
| 566 |
+
"sub-verse076_23": 1,
|
| 567 |
+
"sub-verse076_24": 0,
|
| 568 |
+
"sub-verse082_8": 0,
|
| 569 |
+
"sub-verse082_9": 0,
|
| 570 |
+
"sub-verse082_10": 1,
|
| 571 |
+
"sub-verse082_11": 0,
|
| 572 |
+
"sub-verse082_12": 1,
|
| 573 |
+
"sub-verse082_13": 0,
|
| 574 |
+
"sub-verse082_14": 0,
|
| 575 |
+
"sub-verse082_15": 0,
|
| 576 |
+
"sub-verse082_16": 0,
|
| 577 |
+
"sub-verse082_17": 1,
|
| 578 |
+
"sub-verse082_18": 0,
|
| 579 |
+
"sub-verse082_19": 3,
|
| 580 |
+
"sub-verse082_20": 2,
|
| 581 |
+
"sub-verse082_21": 1,
|
| 582 |
+
"sub-verse082_22": 1,
|
| 583 |
+
"sub-verse082_23": 3,
|
| 584 |
+
"sub-verse082_24": 2,
|
| 585 |
+
"sub-verse088_16": 0,
|
| 586 |
+
"sub-verse088_17": 0,
|
| 587 |
+
"sub-verse088_18": 0,
|
| 588 |
+
"sub-verse088_19": 0,
|
| 589 |
+
"sub-verse088_20": 0,
|
| 590 |
+
"sub-verse088_21": 0,
|
| 591 |
+
"sub-verse088_22": 0,
|
| 592 |
+
"sub-verse088_23": 0,
|
| 593 |
+
"sub-verse088_24": 0,
|
| 594 |
+
"sub-verse091_8": 0,
|
| 595 |
+
"sub-verse091_9": 0,
|
| 596 |
+
"sub-verse091_10": 0,
|
| 597 |
+
"sub-verse091_11": 0,
|
| 598 |
+
"sub-verse091_12": 0,
|
| 599 |
+
"sub-verse091_13": 0,
|
| 600 |
+
"sub-verse091_14": 0,
|
| 601 |
+
"sub-verse091_15": 1,
|
| 602 |
+
"sub-verse091_16": 0,
|
| 603 |
+
"sub-verse091_17": 0,
|
| 604 |
+
"sub-verse091_18": 0,
|
| 605 |
+
"sub-verse091_19": 1,
|
| 606 |
+
"sub-verse091_20": 1,
|
| 607 |
+
"sub-verse091_21": 0,
|
| 608 |
+
"sub-verse091_22": 1,
|
| 609 |
+
"sub-verse091_23": 1,
|
| 610 |
+
"sub-verse091_24": 0,
|
| 611 |
+
"sub-verse096_8": 0,
|
| 612 |
+
"sub-verse096_9": 0,
|
| 613 |
+
"sub-verse096_10": 0,
|
| 614 |
+
"sub-verse096_11": 0,
|
| 615 |
+
"sub-verse096_12": 0,
|
| 616 |
+
"sub-verse096_13": 0,
|
| 617 |
+
"sub-verse096_14": 0,
|
| 618 |
+
"sub-verse096_15": 0,
|
| 619 |
+
"sub-verse096_16": 0,
|
| 620 |
+
"sub-verse096_17": 0,
|
| 621 |
+
"sub-verse096_18": 0,
|
| 622 |
+
"sub-verse096_19": 0,
|
| 623 |
+
"sub-verse096_20": 0,
|
| 624 |
+
"sub-verse096_21": 0,
|
| 625 |
+
"sub-verse096_22": 0,
|
| 626 |
+
"sub-verse096_23": 0,
|
| 627 |
+
"sub-verse096_24": 0,
|
| 628 |
+
"sub-verse097_8": 0,
|
| 629 |
+
"sub-verse097_9": 1,
|
| 630 |
+
"sub-verse097_10": 3,
|
| 631 |
+
"sub-verse097_11": 1,
|
| 632 |
+
"sub-verse097_12": 1,
|
| 633 |
+
"sub-verse097_13": 1,
|
| 634 |
+
"sub-verse097_14": 1,
|
| 635 |
+
"sub-verse097_15": 1,
|
| 636 |
+
"sub-verse097_16": 1,
|
| 637 |
+
"sub-verse097_17": 1,
|
| 638 |
+
"sub-verse097_18": 0,
|
| 639 |
+
"sub-verse097_19": 0,
|
| 640 |
+
"sub-verse097_20": 2,
|
| 641 |
+
"sub-verse097_21": 1,
|
| 642 |
+
"sub-verse097_22": 2,
|
| 643 |
+
"sub-verse097_23": 0,
|
| 644 |
+
"sub-verse097_24": 0,
|
| 645 |
+
"sub-verse100_16": 0,
|
| 646 |
+
"sub-verse100_17": 0,
|
| 647 |
+
"sub-verse100_18": 0,
|
| 648 |
+
"sub-verse100_19": 0,
|
| 649 |
+
"sub-verse100_20": 0,
|
| 650 |
+
"sub-verse100_21": 0,
|
| 651 |
+
"sub-verse100_22": 0,
|
| 652 |
+
"sub-verse100_23": 0,
|
| 653 |
+
"sub-verse102_20": 0,
|
| 654 |
+
"sub-verse102_21": 0,
|
| 655 |
+
"sub-verse102_22": 2,
|
| 656 |
+
"sub-verse102_23": 0,
|
| 657 |
+
"sub-verse102_24": 0,
|
| 658 |
+
"sub-verse104_9": 0,
|
| 659 |
+
"sub-verse104_10": 0,
|
| 660 |
+
"sub-verse104_11": 0,
|
| 661 |
+
"sub-verse104_12": 0,
|
| 662 |
+
"sub-verse104_13": 0,
|
| 663 |
+
"sub-verse104_14": 0,
|
| 664 |
+
"sub-verse104_15": 0,
|
| 665 |
+
"sub-verse104_16": 0,
|
| 666 |
+
"sub-verse104_17": 0,
|
| 667 |
+
"sub-verse104_18": 0,
|
| 668 |
+
"sub-verse104_19": 2,
|
| 669 |
+
"sub-verse104_20": 0,
|
| 670 |
+
"sub-verse104_21": 2,
|
| 671 |
+
"sub-verse104_22": 1,
|
| 672 |
+
"sub-verse104_23": 2,
|
| 673 |
+
"sub-verse104_24": 0,
|
| 674 |
+
"sub-verse105_9": 0,
|
| 675 |
+
"sub-verse105_10": 0,
|
| 676 |
+
"sub-verse105_11": 0,
|
| 677 |
+
"sub-verse105_12": 0,
|
| 678 |
+
"sub-verse105_13": 2,
|
| 679 |
+
"sub-verse105_14": 3,
|
| 680 |
+
"sub-verse105_15": 2,
|
| 681 |
+
"sub-verse105_16": 0,
|
| 682 |
+
"sub-verse105_17": 0,
|
| 683 |
+
"sub-verse107_19": 0,
|
| 684 |
+
"sub-verse107_20": 0,
|
| 685 |
+
"sub-verse107_21": 0,
|
| 686 |
+
"sub-verse107_22": 0,
|
| 687 |
+
"sub-verse107_23": 0,
|
| 688 |
+
"sub-verse107_24": 0,
|
| 689 |
+
"sub-verse111_18": 0,
|
| 690 |
+
"sub-verse111_19": 0,
|
| 691 |
+
"sub-verse111_20": 0,
|
| 692 |
+
"sub-verse111_21": 2,
|
| 693 |
+
"sub-verse111_22": 3,
|
| 694 |
+
"sub-verse111_23": 2,
|
| 695 |
+
"sub-verse111_24": 0,
|
| 696 |
+
"sub-verse112_8": 0,
|
| 697 |
+
"sub-verse112_9": 0,
|
| 698 |
+
"sub-verse112_10": 0,
|
| 699 |
+
"sub-verse112_11": 0,
|
| 700 |
+
"sub-verse112_12": 0,
|
| 701 |
+
"sub-verse112_13": 0,
|
| 702 |
+
"sub-verse112_14": 0,
|
| 703 |
+
"sub-verse112_15": 0,
|
| 704 |
+
"sub-verse112_16": 0,
|
| 705 |
+
"sub-verse112_17": 0,
|
| 706 |
+
"sub-verse112_18": 0,
|
| 707 |
+
"sub-verse112_19": 0,
|
| 708 |
+
"sub-verse112_20": 0,
|
| 709 |
+
"sub-verse112_21": 0,
|
| 710 |
+
"sub-verse112_22": 0,
|
| 711 |
+
"sub-verse112_23": 0,
|
| 712 |
+
"sub-verse112_24": 0,
|
| 713 |
+
"sub-verse113_21": 0,
|
| 714 |
+
"sub-verse113_22": 0,
|
| 715 |
+
"sub-verse113_23": 1,
|
| 716 |
+
"sub-verse113_24": 3,
|
| 717 |
+
"sub-verse113_25": 0,
|
| 718 |
+
"sub-verse122_8": 0,
|
| 719 |
+
"sub-verse122_9": 0,
|
| 720 |
+
"sub-verse122_10": 0,
|
| 721 |
+
"sub-verse122_11": 0,
|
| 722 |
+
"sub-verse122_12": 1,
|
| 723 |
+
"sub-verse122_13": 2,
|
| 724 |
+
"sub-verse122_14": 3,
|
| 725 |
+
"sub-verse122_15": 0,
|
| 726 |
+
"sub-verse122_16": 0,
|
| 727 |
+
"sub-verse122_17": 0,
|
| 728 |
+
"sub-verse122_19": 0,
|
| 729 |
+
"sub-verse122_20": 3,
|
| 730 |
+
"sub-verse122_21": 2,
|
| 731 |
+
"sub-verse122_22": 1,
|
| 732 |
+
"sub-verse122_23": 2,
|
| 733 |
+
"sub-verse122_24": 0,
|
| 734 |
+
"sub-verse127_8": 0,
|
| 735 |
+
"sub-verse127_9": 0,
|
| 736 |
+
"sub-verse127_10": 0,
|
| 737 |
+
"sub-verse127_11": 0,
|
| 738 |
+
"sub-verse127_12": 0,
|
| 739 |
+
"sub-verse127_13": 0,
|
| 740 |
+
"sub-verse127_14": 0,
|
| 741 |
+
"sub-verse127_15": 0,
|
| 742 |
+
"sub-verse127_16": 0,
|
| 743 |
+
"sub-verse127_17": 0,
|
| 744 |
+
"sub-verse127_18": 2,
|
| 745 |
+
"sub-verse127_19": 0,
|
| 746 |
+
"sub-verse127_20": 0,
|
| 747 |
+
"sub-verse127_21": 0,
|
| 748 |
+
"sub-verse127_22": 0,
|
| 749 |
+
"sub-verse127_23": 0,
|
| 750 |
+
"sub-verse127_24": 0,
|
| 751 |
+
"sub-verse133_16": 0,
|
| 752 |
+
"sub-verse133_17": 0,
|
| 753 |
+
"sub-verse133_18": 1,
|
| 754 |
+
"sub-verse133_19": 0,
|
| 755 |
+
"sub-verse133_20": 0,
|
| 756 |
+
"sub-verse133_21": 0,
|
| 757 |
+
"sub-verse133_22": 0,
|
| 758 |
+
"sub-verse133_23": 0,
|
| 759 |
+
"sub-verse133_24": 0,
|
| 760 |
+
"sub-verse134_19": 0,
|
| 761 |
+
"sub-verse134_20": 0,
|
| 762 |
+
"sub-verse134_21": 0,
|
| 763 |
+
"sub-verse134_22": 0,
|
| 764 |
+
"sub-verse134_23": 0,
|
| 765 |
+
"sub-verse134_24": 0,
|
| 766 |
+
"sub-verse135_8": 0,
|
| 767 |
+
"sub-verse135_9": 0,
|
| 768 |
+
"sub-verse135_10": 1,
|
| 769 |
+
"sub-verse135_11": 0,
|
| 770 |
+
"sub-verse135_12": 1,
|
| 771 |
+
"sub-verse135_13": 0,
|
| 772 |
+
"sub-verse135_14": 0,
|
| 773 |
+
"sub-verse135_15": 0,
|
| 774 |
+
"sub-verse135_16": 0,
|
| 775 |
+
"sub-verse135_17": 0,
|
| 776 |
+
"sub-verse135_18": 0,
|
| 777 |
+
"sub-verse135_19": 0,
|
| 778 |
+
"sub-verse135_20": 0,
|
| 779 |
+
"sub-verse135_21": 0,
|
| 780 |
+
"sub-verse135_22": 2,
|
| 781 |
+
"sub-verse135_23": 0,
|
| 782 |
+
"sub-verse135_24": 2,
|
| 783 |
+
"sub-verse137_19": 0,
|
| 784 |
+
"sub-verse137_20": 0,
|
| 785 |
+
"sub-verse137_21": 0,
|
| 786 |
+
"sub-verse137_22": 0,
|
| 787 |
+
"sub-verse137_24": 3,
|
| 788 |
+
"sub-verse139_20": 0,
|
| 789 |
+
"sub-verse139_21": 0,
|
| 790 |
+
"sub-verse139_22": 0,
|
| 791 |
+
"sub-verse139_23": 0,
|
| 792 |
+
"sub-verse139_24": 2,
|
| 793 |
+
"sub-verse141_8": 0,
|
| 794 |
+
"sub-verse141_9": 0,
|
| 795 |
+
"sub-verse141_10": 0,
|
| 796 |
+
"sub-verse141_11": 0,
|
| 797 |
+
"sub-verse141_12": 0,
|
| 798 |
+
"sub-verse141_13": 0,
|
| 799 |
+
"sub-verse141_14": 0,
|
| 800 |
+
"sub-verse141_15": 0,
|
| 801 |
+
"sub-verse141_16": 0,
|
| 802 |
+
"sub-verse141_17": 0,
|
| 803 |
+
"sub-verse141_18": 2,
|
| 804 |
+
"sub-verse141_19": 0,
|
| 805 |
+
"sub-verse141_20": 0,
|
| 806 |
+
"sub-verse141_21": 0,
|
| 807 |
+
"sub-verse141_22": 0,
|
| 808 |
+
"sub-verse141_23": 0,
|
| 809 |
+
"sub-verse141_24": 0,
|
| 810 |
+
"sub-verse145_8": 1,
|
| 811 |
+
"sub-verse145_9": 0,
|
| 812 |
+
"sub-verse145_10": 0,
|
| 813 |
+
"sub-verse145_11": 0,
|
| 814 |
+
"sub-verse145_12": 0,
|
| 815 |
+
"sub-verse145_13": 0,
|
| 816 |
+
"sub-verse145_14": 1,
|
| 817 |
+
"sub-verse145_15": 0,
|
| 818 |
+
"sub-verse145_16": 0,
|
| 819 |
+
"sub-verse145_17": 0,
|
| 820 |
+
"sub-verse145_18": 0,
|
| 821 |
+
"sub-verse145_19": 0,
|
| 822 |
+
"sub-verse145_20": 0,
|
| 823 |
+
"sub-verse145_21": 0,
|
| 824 |
+
"sub-verse145_22": 0,
|
| 825 |
+
"sub-verse145_23": 0,
|
| 826 |
+
"sub-verse145_24": 0,
|
| 827 |
+
"sub-verse146_22": 0,
|
| 828 |
+
"sub-verse146_23": 0,
|
| 829 |
+
"sub-verse151_9": 0,
|
| 830 |
+
"sub-verse151_10": 0,
|
| 831 |
+
"sub-verse151_11": 0,
|
| 832 |
+
"sub-verse151_12": 0,
|
| 833 |
+
"sub-verse151_13": 0,
|
| 834 |
+
"sub-verse151_14": 0,
|
| 835 |
+
"sub-verse151_15": 0,
|
| 836 |
+
"sub-verse151_16": 0,
|
| 837 |
+
"sub-verse151_17": 0,
|
| 838 |
+
"sub-verse151_18": 0,
|
| 839 |
+
"sub-verse151_19": 2,
|
| 840 |
+
"sub-verse151_20": 0,
|
| 841 |
+
"sub-verse151_21": 0,
|
| 842 |
+
"sub-verse151_22": 0,
|
| 843 |
+
"sub-verse151_23": 0,
|
| 844 |
+
"sub-verse151_24": 0,
|
| 845 |
+
"sub-verse152_19": 1,
|
| 846 |
+
"sub-verse152_20": 2,
|
| 847 |
+
"sub-verse152_21": 1,
|
| 848 |
+
"sub-verse152_22": 1,
|
| 849 |
+
"sub-verse152_23": 1,
|
| 850 |
+
"sub-verse152_24": 0,
|
| 851 |
+
"sub-verse401_series0_8": 0,
|
| 852 |
+
"sub-verse401_series0_9": 0,
|
| 853 |
+
"sub-verse402_series0_8": 0,
|
| 854 |
+
"sub-verse207_8": 0,
|
| 855 |
+
"sub-verse403_series0_8": 0,
|
| 856 |
+
"sub-verse403_series0_9": 0,
|
| 857 |
+
"sub-verse405_series0_8": 0,
|
| 858 |
+
"sub-verse405_series0_9": 0,
|
| 859 |
+
"sub-verse406_series0_8": 0,
|
| 860 |
+
"sub-verse406_series0_9": 0,
|
| 861 |
+
"sub-verse408_series0_8": 0,
|
| 862 |
+
"sub-verse409_series0_8": 0,
|
| 863 |
+
"sub-verse409_series0_9": 0,
|
| 864 |
+
"sub-verse410_series0_8": 0,
|
| 865 |
+
"sub-verse411_series0_8": 0,
|
| 866 |
+
"sub-verse413_series0_8": 0,
|
| 867 |
+
"sub-verse413_series0_9": 0,
|
| 868 |
+
"sub-verse413_series0_10": 0,
|
| 869 |
+
"sub-verse413_series0_11": 0,
|
| 870 |
+
"sub-verse415_series0_8": 0,
|
| 871 |
+
"sub-verse402_series1_8": 0,
|
| 872 |
+
"sub-verse402_series1_9": 0,
|
| 873 |
+
"sub-verse402_series1_10": 0,
|
| 874 |
+
"sub-verse402_series1_11": 0,
|
| 875 |
+
"sub-verse402_series1_12": 0,
|
| 876 |
+
"sub-verse402_series1_13": 0,
|
| 877 |
+
"sub-verse402_series1_14": 0,
|
| 878 |
+
"sub-verse402_series1_15": 0,
|
| 879 |
+
"sub-verse402_series1_16": 0,
|
| 880 |
+
"sub-verse402_series1_17": 0,
|
| 881 |
+
"sub-verse402_series1_18": 0,
|
| 882 |
+
"sub-verse402_series1_19": 0,
|
| 883 |
+
"sub-verse402_series1_20": 0,
|
| 884 |
+
"sub-verse402_series1_21": 0,
|
| 885 |
+
"sub-verse402_series1_22": 0,
|
| 886 |
+
"sub-verse402_series1_23": 0,
|
| 887 |
+
"sub-verse402_series1_24": 0,
|
| 888 |
+
"sub-verse401_series1_10": 0,
|
| 889 |
+
"sub-verse401_series1_11": 0,
|
| 890 |
+
"sub-verse401_series1_12": 0,
|
| 891 |
+
"sub-verse401_series1_13": 0,
|
| 892 |
+
"sub-verse401_series1_14": 0,
|
| 893 |
+
"sub-verse401_series1_15": 0,
|
| 894 |
+
"sub-verse401_series1_16": 0,
|
| 895 |
+
"sub-verse401_series1_17": 0,
|
| 896 |
+
"sub-verse401_series1_18": 1,
|
| 897 |
+
"sub-verse401_series1_19": 1,
|
| 898 |
+
"sub-verse401_series1_20": 0,
|
| 899 |
+
"sub-verse401_series1_21": 0,
|
| 900 |
+
"sub-verse401_series1_22": 0,
|
| 901 |
+
"sub-verse401_series1_23": 0,
|
| 902 |
+
"sub-verse401_series1_24": 0,
|
| 903 |
+
"sub-verse254_8": 0,
|
| 904 |
+
"sub-verse254_9": 0,
|
| 905 |
+
"sub-verse254_10": 0,
|
| 906 |
+
"sub-verse254_11": 0,
|
| 907 |
+
"sub-verse254_12": 0,
|
| 908 |
+
"sub-verse254_13": 0,
|
| 909 |
+
"sub-verse254_14": 0,
|
| 910 |
+
"sub-verse254_15": 0,
|
| 911 |
+
"sub-verse254_16": 0,
|
| 912 |
+
"sub-verse254_17": 0,
|
| 913 |
+
"sub-verse254_18": 0,
|
| 914 |
+
"sub-verse254_19": 0,
|
| 915 |
+
"sub-verse254_20": 0,
|
| 916 |
+
"sub-verse254_21": 0,
|
| 917 |
+
"sub-verse254_22": 0,
|
| 918 |
+
"sub-verse254_23": 0,
|
| 919 |
+
"sub-verse254_24": 0,
|
| 920 |
+
"sub-verse403_series1_8": 0,
|
| 921 |
+
"sub-verse403_series1_9": 0,
|
| 922 |
+
"sub-verse403_series1_10": 0,
|
| 923 |
+
"sub-verse403_series1_11": 0,
|
| 924 |
+
"sub-verse403_series1_12": 0,
|
| 925 |
+
"sub-verse403_series1_13": 1,
|
| 926 |
+
"sub-verse403_series1_14": 1,
|
| 927 |
+
"sub-verse403_series1_15": 1,
|
| 928 |
+
"sub-verse403_series1_16": 1,
|
| 929 |
+
"sub-verse403_series1_17": 0,
|
| 930 |
+
"sub-verse403_series1_18": 0,
|
| 931 |
+
"sub-verse403_series1_19": 0,
|
| 932 |
+
"sub-verse403_series1_20": 0,
|
| 933 |
+
"sub-verse403_series1_21": 0,
|
| 934 |
+
"sub-verse403_series1_22": 0,
|
| 935 |
+
"sub-verse403_series1_23": 0,
|
| 936 |
+
"sub-verse403_series1_24": 0,
|
| 937 |
+
"sub-verse403_series1_25": 0,
|
| 938 |
+
"sub-verse257_17": 0,
|
| 939 |
+
"sub-verse257_18": 0,
|
| 940 |
+
"sub-verse257_19": 0,
|
| 941 |
+
"sub-verse257_20": 0,
|
| 942 |
+
"sub-verse257_21": 0,
|
| 943 |
+
"sub-verse257_22": 0,
|
| 944 |
+
"sub-verse257_23": 0,
|
| 945 |
+
"sub-verse257_24": 0,
|
| 946 |
+
"sub-verse405_series1_8": 0,
|
| 947 |
+
"sub-verse405_series1_9": 0,
|
| 948 |
+
"sub-verse405_series1_10": 0,
|
| 949 |
+
"sub-verse405_series1_11": 0,
|
| 950 |
+
"sub-verse405_series1_12": 0,
|
| 951 |
+
"sub-verse405_series1_13": 0,
|
| 952 |
+
"sub-verse405_series1_14": 0,
|
| 953 |
+
"sub-verse405_series1_15": 0,
|
| 954 |
+
"sub-verse405_series1_16": 0,
|
| 955 |
+
"sub-verse405_series1_17": 0,
|
| 956 |
+
"sub-verse405_series1_18": 0,
|
| 957 |
+
"sub-verse405_series1_19": 0,
|
| 958 |
+
"sub-verse405_series2_20": 1,
|
| 959 |
+
"sub-verse405_series2_21": 2,
|
| 960 |
+
"sub-verse405_series2_22": 0,
|
| 961 |
+
"sub-verse405_series2_23": 0,
|
| 962 |
+
"sub-verse405_series2_24": 0,
|
| 963 |
+
"sub-verse406_series1_8": 0,
|
| 964 |
+
"sub-verse406_series1_9": 0,
|
| 965 |
+
"sub-verse406_series1_10": 0,
|
| 966 |
+
"sub-verse406_series1_11": 0,
|
| 967 |
+
"sub-verse406_series1_12": 0,
|
| 968 |
+
"sub-verse406_series1_13": 0,
|
| 969 |
+
"sub-verse406_series1_14": 0,
|
| 970 |
+
"sub-verse406_series1_15": 0,
|
| 971 |
+
"sub-verse406_series1_16": 0,
|
| 972 |
+
"sub-verse406_series1_17": 0,
|
| 973 |
+
"sub-verse406_series1_18": 0,
|
| 974 |
+
"sub-verse406_series1_19": 0,
|
| 975 |
+
"sub-verse406_series1_20": 0,
|
| 976 |
+
"sub-verse406_series1_21": 0,
|
| 977 |
+
"sub-verse406_series1_22": 0,
|
| 978 |
+
"sub-verse406_series1_23": 0,
|
| 979 |
+
"sub-verse406_series1_24": 0,
|
| 980 |
+
"sub-verse406_series1_25": 0,
|
| 981 |
+
"sub-verse407_series1_8": 0,
|
| 982 |
+
"sub-verse407_series1_9": 0,
|
| 983 |
+
"sub-verse407_series1_10": 0,
|
| 984 |
+
"sub-verse407_series1_11": 0,
|
| 985 |
+
"sub-verse407_series1_12": 0,
|
| 986 |
+
"sub-verse407_series1_13": 0,
|
| 987 |
+
"sub-verse407_series1_14": 0,
|
| 988 |
+
"sub-verse407_series1_15": 0,
|
| 989 |
+
"sub-verse407_series1_16": 0,
|
| 990 |
+
"sub-verse407_series1_17": 0,
|
| 991 |
+
"sub-verse407_series1_18": 0,
|
| 992 |
+
"sub-verse407_series1_19": 0,
|
| 993 |
+
"sub-verse407_series1_20": 0,
|
| 994 |
+
"sub-verse407_series1_21": 0,
|
| 995 |
+
"sub-verse407_series1_22": 0,
|
| 996 |
+
"sub-verse407_series1_23": 0,
|
| 997 |
+
"sub-verse407_series1_24": 0,
|
| 998 |
+
"sub-verse408_series1_8": 0,
|
| 999 |
+
"sub-verse408_series1_9": 0,
|
| 1000 |
+
"sub-verse408_series1_10": 0,
|
| 1001 |
+
"sub-verse408_series1_11": 0,
|
| 1002 |
+
"sub-verse408_series1_12": 0,
|
| 1003 |
+
"sub-verse408_series1_13": 0,
|
| 1004 |
+
"sub-verse408_series1_14": 0,
|
| 1005 |
+
"sub-verse408_series1_15": 0,
|
| 1006 |
+
"sub-verse408_series1_16": 0,
|
| 1007 |
+
"sub-verse408_series1_17": 0,
|
| 1008 |
+
"sub-verse408_series1_18": 0,
|
| 1009 |
+
"sub-verse408_series1_19": 2,
|
| 1010 |
+
"sub-verse408_series1_20": 3,
|
| 1011 |
+
"sub-verse408_series1_21": 1,
|
| 1012 |
+
"sub-verse408_series1_22": 0,
|
| 1013 |
+
"sub-verse408_series1_23": 2,
|
| 1014 |
+
"sub-verse408_series1_24": 0,
|
| 1015 |
+
"sub-verse409_series1_8": 0,
|
| 1016 |
+
"sub-verse409_series1_9": 0,
|
| 1017 |
+
"sub-verse409_series1_10": 0,
|
| 1018 |
+
"sub-verse409_series1_11": 0,
|
| 1019 |
+
"sub-verse409_series1_12": 0,
|
| 1020 |
+
"sub-verse409_series1_13": 0,
|
| 1021 |
+
"sub-verse409_series1_14": 0,
|
| 1022 |
+
"sub-verse409_series1_15": 0,
|
| 1023 |
+
"sub-verse409_series1_16": 0,
|
| 1024 |
+
"sub-verse409_series1_17": 0,
|
| 1025 |
+
"sub-verse409_series1_18": 0,
|
| 1026 |
+
"sub-verse409_series1_19": 2,
|
| 1027 |
+
"sub-verse409_series1_20": 2,
|
| 1028 |
+
"sub-verse409_series1_21": 3,
|
| 1029 |
+
"sub-verse409_series1_22": 3,
|
| 1030 |
+
"sub-verse409_series1_23": 3,
|
| 1031 |
+
"sub-verse409_series1_24": 1,
|
| 1032 |
+
"sub-verse410_series1_8": 0,
|
| 1033 |
+
"sub-verse410_series1_9": 0,
|
| 1034 |
+
"sub-verse410_series1_10": 0,
|
| 1035 |
+
"sub-verse410_series1_11": 0,
|
| 1036 |
+
"sub-verse410_series1_12": 0,
|
| 1037 |
+
"sub-verse410_series1_13": 0,
|
| 1038 |
+
"sub-verse410_series1_14": 0,
|
| 1039 |
+
"sub-verse410_series1_15": 0,
|
| 1040 |
+
"sub-verse410_series1_16": 0,
|
| 1041 |
+
"sub-verse410_series1_17": 0,
|
| 1042 |
+
"sub-verse410_series1_18": 0,
|
| 1043 |
+
"sub-verse410_series1_19": 1,
|
| 1044 |
+
"sub-verse410_series1_20": 1,
|
| 1045 |
+
"sub-verse410_series1_21": 2,
|
| 1046 |
+
"sub-verse410_series1_22": 0,
|
| 1047 |
+
"sub-verse410_series1_23": 0,
|
| 1048 |
+
"sub-verse410_series1_24": 0,
|
| 1049 |
+
"sub-verse411_series1_8": 0,
|
| 1050 |
+
"sub-verse411_series1_9": 0,
|
| 1051 |
+
"sub-verse411_series1_10": 0,
|
| 1052 |
+
"sub-verse411_series1_11": 0,
|
| 1053 |
+
"sub-verse411_series1_12": 0,
|
| 1054 |
+
"sub-verse411_series1_13": 0,
|
| 1055 |
+
"sub-verse411_series1_14": 0,
|
| 1056 |
+
"sub-verse411_series1_15": 0,
|
| 1057 |
+
"sub-verse411_series1_16": 0,
|
| 1058 |
+
"sub-verse411_series1_17": 0,
|
| 1059 |
+
"sub-verse411_series1_18": 0,
|
| 1060 |
+
"sub-verse411_series1_19": 0,
|
| 1061 |
+
"sub-verse411_series1_20": 0,
|
| 1062 |
+
"sub-verse411_series1_21": 0,
|
| 1063 |
+
"sub-verse411_series1_22": 0,
|
| 1064 |
+
"sub-verse411_series1_23": 0,
|
| 1065 |
+
"sub-verse411_series1_24": 0,
|
| 1066 |
+
"sub-verse413_series1_8": 0,
|
| 1067 |
+
"sub-verse413_series1_9": 0,
|
| 1068 |
+
"sub-verse413_series1_10": 0,
|
| 1069 |
+
"sub-verse413_series1_11": 0,
|
| 1070 |
+
"sub-verse413_series1_12": 0,
|
| 1071 |
+
"sub-verse413_series1_13": 0,
|
| 1072 |
+
"sub-verse413_series1_14": 0,
|
| 1073 |
+
"sub-verse413_series1_15": 0,
|
| 1074 |
+
"sub-verse413_series1_16": 0,
|
| 1075 |
+
"sub-verse413_series1_17": 0,
|
| 1076 |
+
"sub-verse413_series1_18": 0,
|
| 1077 |
+
"sub-verse413_series1_19": 0,
|
| 1078 |
+
"sub-verse413_series1_20": 0,
|
| 1079 |
+
"sub-verse413_series1_21": 0,
|
| 1080 |
+
"sub-verse413_series1_22": 0,
|
| 1081 |
+
"sub-verse413_series1_23": 0,
|
| 1082 |
+
"sub-verse413_series1_24": 0,
|
| 1083 |
+
"sub-verse415_series1_8": 0,
|
| 1084 |
+
"sub-verse415_series1_9": 0,
|
| 1085 |
+
"sub-verse415_series1_10": 0,
|
| 1086 |
+
"sub-verse415_series1_11": 0,
|
| 1087 |
+
"sub-verse415_series1_12": 0,
|
| 1088 |
+
"sub-verse415_series1_13": 0,
|
| 1089 |
+
"sub-verse415_series1_14": 0,
|
| 1090 |
+
"sub-verse415_series1_15": 0,
|
| 1091 |
+
"sub-verse415_series1_16": 0,
|
| 1092 |
+
"sub-verse415_series1_17": 0,
|
| 1093 |
+
"sub-verse415_series1_18": 0,
|
| 1094 |
+
"sub-verse415_series1_19": 0,
|
| 1095 |
+
"sub-verse415_series1_20": 0,
|
| 1096 |
+
"sub-verse415_series1_21": 1,
|
| 1097 |
+
"sub-verse415_series1_22": 0,
|
| 1098 |
+
"sub-verse415_series1_23": 0,
|
| 1099 |
+
"sub-verse415_series1_24": 0
|
| 1100 |
+
},
|
| 1101 |
+
"val": {
|
| 1102 |
+
"sub-verse010_20": 3,
|
| 1103 |
+
"sub-verse010_21": 0,
|
| 1104 |
+
"sub-verse010_22": 0,
|
| 1105 |
+
"sub-verse010_23": 0,
|
| 1106 |
+
"sub-verse010_24": 0,
|
| 1107 |
+
"sub-verse011_8": 1,
|
| 1108 |
+
"sub-verse011_9": 0,
|
| 1109 |
+
"sub-verse011_10": 1,
|
| 1110 |
+
"sub-verse011_11": 0,
|
| 1111 |
+
"sub-verse011_12": 0,
|
| 1112 |
+
"sub-verse011_13": 3,
|
| 1113 |
+
"sub-verse011_14": 1,
|
| 1114 |
+
"sub-verse011_15": 3,
|
| 1115 |
+
"sub-verse011_16": 0,
|
| 1116 |
+
"sub-verse011_17": 0,
|
| 1117 |
+
"sub-verse011_18": 0,
|
| 1118 |
+
"sub-verse011_19": 0,
|
| 1119 |
+
"sub-verse013_9": 0,
|
| 1120 |
+
"sub-verse013_10": 0,
|
| 1121 |
+
"sub-verse013_11": 0,
|
| 1122 |
+
"sub-verse013_12": 0,
|
| 1123 |
+
"sub-verse013_13": 0,
|
| 1124 |
+
"sub-verse013_14": 0,
|
| 1125 |
+
"sub-verse013_15": 0,
|
| 1126 |
+
"sub-verse013_16": 0,
|
| 1127 |
+
"sub-verse013_17": 0,
|
| 1128 |
+
"sub-verse013_18": 0,
|
| 1129 |
+
"sub-verse013_19": 0,
|
| 1130 |
+
"sub-verse013_20": 3,
|
| 1131 |
+
"sub-verse013_21": 2,
|
| 1132 |
+
"sub-verse013_22": 1,
|
| 1133 |
+
"sub-verse013_23": 1,
|
| 1134 |
+
"sub-verse013_24": 1,
|
| 1135 |
+
"sub-verse016_8": 0,
|
| 1136 |
+
"sub-verse016_9": 0,
|
| 1137 |
+
"sub-verse016_10": 0,
|
| 1138 |
+
"sub-verse016_11": 0,
|
| 1139 |
+
"sub-verse016_12": 0,
|
| 1140 |
+
"sub-verse016_13": 0,
|
| 1141 |
+
"sub-verse016_14": 1,
|
| 1142 |
+
"sub-verse016_15": 0,
|
| 1143 |
+
"sub-verse016_16": 0,
|
| 1144 |
+
"sub-verse016_17": 0,
|
| 1145 |
+
"sub-verse016_18": 0,
|
| 1146 |
+
"sub-verse016_19": 0,
|
| 1147 |
+
"sub-verse016_20": 0,
|
| 1148 |
+
"sub-verse016_21": 0,
|
| 1149 |
+
"sub-verse016_22": 0,
|
| 1150 |
+
"sub-verse016_23": 0,
|
| 1151 |
+
"sub-verse016_24": 0,
|
| 1152 |
+
"sub-verse018_8": 0,
|
| 1153 |
+
"sub-verse018_9": 0,
|
| 1154 |
+
"sub-verse018_10": 0,
|
| 1155 |
+
"sub-verse018_11": 0,
|
| 1156 |
+
"sub-verse018_12": 0,
|
| 1157 |
+
"sub-verse018_13": 0,
|
| 1158 |
+
"sub-verse018_14": 0,
|
| 1159 |
+
"sub-verse018_15": 0,
|
| 1160 |
+
"sub-verse018_16": 0,
|
| 1161 |
+
"sub-verse018_17": 0,
|
| 1162 |
+
"sub-verse018_18": 0,
|
| 1163 |
+
"sub-verse018_19": 0,
|
| 1164 |
+
"sub-verse018_20": 0,
|
| 1165 |
+
"sub-verse018_21": 0,
|
| 1166 |
+
"sub-verse018_22": 0,
|
| 1167 |
+
"sub-verse018_23": 0,
|
| 1168 |
+
"sub-verse022_20": 0,
|
| 1169 |
+
"sub-verse022_21": 0,
|
| 1170 |
+
"sub-verse022_22": 3,
|
| 1171 |
+
"sub-verse022_23": 2,
|
| 1172 |
+
"sub-verse022_24": 0,
|
| 1173 |
+
"sub-verse023_18": 0,
|
| 1174 |
+
"sub-verse023_19": 0,
|
| 1175 |
+
"sub-verse023_20": 1,
|
| 1176 |
+
"sub-verse023_21": 0,
|
| 1177 |
+
"sub-verse023_22": 0,
|
| 1178 |
+
"sub-verse023_23": 0,
|
| 1179 |
+
"sub-verse023_24": 1,
|
| 1180 |
+
"sub-verse023_25": 0,
|
| 1181 |
+
"sub-verse024_15": 0,
|
| 1182 |
+
"sub-verse024_16": 0,
|
| 1183 |
+
"sub-verse024_17": 0,
|
| 1184 |
+
"sub-verse024_18": 0,
|
| 1185 |
+
"sub-verse024_19": 1,
|
| 1186 |
+
"sub-verse024_20": 3,
|
| 1187 |
+
"sub-verse024_21": 0,
|
| 1188 |
+
"sub-verse024_22": 0,
|
| 1189 |
+
"sub-verse024_23": 0,
|
| 1190 |
+
"sub-verse024_24": 0,
|
| 1191 |
+
"sub-verse026_14": 0,
|
| 1192 |
+
"sub-verse026_15": 1,
|
| 1193 |
+
"sub-verse026_16": 0,
|
| 1194 |
+
"sub-verse026_17": 0,
|
| 1195 |
+
"sub-verse026_18": 0,
|
| 1196 |
+
"sub-verse026_19": 3,
|
| 1197 |
+
"sub-verse026_20": 1,
|
| 1198 |
+
"sub-verse026_21": 0,
|
| 1199 |
+
"sub-verse026_22": 0,
|
| 1200 |
+
"sub-verse026_23": 2,
|
| 1201 |
+
"sub-verse026_24": 2,
|
| 1202 |
+
"sub-verse030_20": 2,
|
| 1203 |
+
"sub-verse030_21": 1,
|
| 1204 |
+
"sub-verse030_22": 1,
|
| 1205 |
+
"sub-verse030_23": 0,
|
| 1206 |
+
"sub-verse030_24": 0,
|
| 1207 |
+
"sub-verse041_8": 0,
|
| 1208 |
+
"sub-verse041_9": 0,
|
| 1209 |
+
"sub-verse041_10": 0,
|
| 1210 |
+
"sub-verse041_11": 0,
|
| 1211 |
+
"sub-verse041_12": 0,
|
| 1212 |
+
"sub-verse041_13": 0,
|
| 1213 |
+
"sub-verse041_14": 0,
|
| 1214 |
+
"sub-verse041_15": 0,
|
| 1215 |
+
"sub-verse041_16": 0,
|
| 1216 |
+
"sub-verse041_17": 0,
|
| 1217 |
+
"sub-verse041_18": 0,
|
| 1218 |
+
"sub-verse041_19": 0,
|
| 1219 |
+
"sub-verse041_20": 0,
|
| 1220 |
+
"sub-verse041_21": 0,
|
| 1221 |
+
"sub-verse041_22": 0,
|
| 1222 |
+
"sub-verse041_23": 0,
|
| 1223 |
+
"sub-verse041_24": 0,
|
| 1224 |
+
"sub-verse047_8": 0,
|
| 1225 |
+
"sub-verse047_9": 0,
|
| 1226 |
+
"sub-verse047_10": 0,
|
| 1227 |
+
"sub-verse047_11": 0,
|
| 1228 |
+
"sub-verse047_12": 0,
|
| 1229 |
+
"sub-verse047_13": 0,
|
| 1230 |
+
"sub-verse047_14": 3,
|
| 1231 |
+
"sub-verse047_15": 0,
|
| 1232 |
+
"sub-verse047_16": 0,
|
| 1233 |
+
"sub-verse047_17": 0,
|
| 1234 |
+
"sub-verse047_18": 1,
|
| 1235 |
+
"sub-verse047_19": 0,
|
| 1236 |
+
"sub-verse047_20": 0,
|
| 1237 |
+
"sub-verse047_21": 0,
|
| 1238 |
+
"sub-verse047_22": 0,
|
| 1239 |
+
"sub-verse047_23": 0,
|
| 1240 |
+
"sub-verse047_24": 0,
|
| 1241 |
+
"sub-verse058_19": 0,
|
| 1242 |
+
"sub-verse058_20": 0,
|
| 1243 |
+
"sub-verse058_21": 0,
|
| 1244 |
+
"sub-verse058_22": 0,
|
| 1245 |
+
"sub-verse058_23": 0,
|
| 1246 |
+
"sub-verse058_24": 0,
|
| 1247 |
+
"sub-verse067_15": 0,
|
| 1248 |
+
"sub-verse067_16": 0,
|
| 1249 |
+
"sub-verse067_17": 0,
|
| 1250 |
+
"sub-verse067_18": 0,
|
| 1251 |
+
"sub-verse067_19": 0,
|
| 1252 |
+
"sub-verse067_20": 0,
|
| 1253 |
+
"sub-verse067_21": 0,
|
| 1254 |
+
"sub-verse067_22": 0,
|
| 1255 |
+
"sub-verse067_23": 0,
|
| 1256 |
+
"sub-verse067_24": 0,
|
| 1257 |
+
"sub-verse071_20": 0,
|
| 1258 |
+
"sub-verse071_21": 0,
|
| 1259 |
+
"sub-verse071_22": 0,
|
| 1260 |
+
"sub-verse071_23": 0,
|
| 1261 |
+
"sub-verse071_24": 0,
|
| 1262 |
+
"sub-verse073_20": 0,
|
| 1263 |
+
"sub-verse073_21": 0,
|
| 1264 |
+
"sub-verse073_22": 0,
|
| 1265 |
+
"sub-verse073_23": 0,
|
| 1266 |
+
"sub-verse073_24": 0,
|
| 1267 |
+
"sub-verse078_19": 0,
|
| 1268 |
+
"sub-verse078_20": 3,
|
| 1269 |
+
"sub-verse078_21": 0,
|
| 1270 |
+
"sub-verse078_22": 0,
|
| 1271 |
+
"sub-verse078_23": 0,
|
| 1272 |
+
"sub-verse078_24": 0,
|
| 1273 |
+
"sub-verse080_11": 2,
|
| 1274 |
+
"sub-verse080_12": 0,
|
| 1275 |
+
"sub-verse080_13": 0,
|
| 1276 |
+
"sub-verse080_14": 1,
|
| 1277 |
+
"sub-verse080_15": 1,
|
| 1278 |
+
"sub-verse080_16": 2,
|
| 1279 |
+
"sub-verse080_17": 0,
|
| 1280 |
+
"sub-verse080_18": 0,
|
| 1281 |
+
"sub-verse080_19": 0,
|
| 1282 |
+
"sub-verse080_20": 2,
|
| 1283 |
+
"sub-verse080_21": 1,
|
| 1284 |
+
"sub-verse080_22": 0,
|
| 1285 |
+
"sub-verse080_23": 2,
|
| 1286 |
+
"sub-verse080_24": 1,
|
| 1287 |
+
"sub-verse093_16": 0,
|
| 1288 |
+
"sub-verse093_17": 0,
|
| 1289 |
+
"sub-verse093_20": 3,
|
| 1290 |
+
"sub-verse093_23": 2,
|
| 1291 |
+
"sub-verse093_24": 0,
|
| 1292 |
+
"sub-verse095_8": 0,
|
| 1293 |
+
"sub-verse095_9": 0,
|
| 1294 |
+
"sub-verse095_10": 0,
|
| 1295 |
+
"sub-verse095_11": 2,
|
| 1296 |
+
"sub-verse095_12": 0,
|
| 1297 |
+
"sub-verse095_13": 0,
|
| 1298 |
+
"sub-verse095_14": 0,
|
| 1299 |
+
"sub-verse095_15": 0,
|
| 1300 |
+
"sub-verse095_16": 0,
|
| 1301 |
+
"sub-verse095_17": 0,
|
| 1302 |
+
"sub-verse095_18": 0,
|
| 1303 |
+
"sub-verse095_19": 0,
|
| 1304 |
+
"sub-verse095_20": 0,
|
| 1305 |
+
"sub-verse095_21": 0,
|
| 1306 |
+
"sub-verse095_22": 1,
|
| 1307 |
+
"sub-verse095_23": 2,
|
| 1308 |
+
"sub-verse095_24": 0,
|
| 1309 |
+
"sub-verse116_24": 0,
|
| 1310 |
+
"sub-verse124_20": 1,
|
| 1311 |
+
"sub-verse124_21": 2,
|
| 1312 |
+
"sub-verse124_22": 2,
|
| 1313 |
+
"sub-verse124_23": 2,
|
| 1314 |
+
"sub-verse124_24": 1,
|
| 1315 |
+
"sub-verse125_9": 0,
|
| 1316 |
+
"sub-verse125_10": 1,
|
| 1317 |
+
"sub-verse125_11": 0,
|
| 1318 |
+
"sub-verse125_12": 0,
|
| 1319 |
+
"sub-verse125_13": 3,
|
| 1320 |
+
"sub-verse125_14": 0,
|
| 1321 |
+
"sub-verse125_15": 0,
|
| 1322 |
+
"sub-verse125_16": 1,
|
| 1323 |
+
"sub-verse125_17": 1,
|
| 1324 |
+
"sub-verse125_18": 1,
|
| 1325 |
+
"sub-verse125_19": 2,
|
| 1326 |
+
"sub-verse125_20": 2,
|
| 1327 |
+
"sub-verse150_16": 0,
|
| 1328 |
+
"sub-verse150_17": 0,
|
| 1329 |
+
"sub-verse150_18": 1,
|
| 1330 |
+
"sub-verse150_20": 3,
|
| 1331 |
+
"sub-verse150_21": 0,
|
| 1332 |
+
"sub-verse153_16": 1,
|
| 1333 |
+
"sub-verse153_17": 2,
|
| 1334 |
+
"sub-verse153_18": 2,
|
| 1335 |
+
"sub-verse153_19": 1,
|
| 1336 |
+
"sub-verse153_20": 3,
|
| 1337 |
+
"sub-verse153_21": 1,
|
| 1338 |
+
"sub-verse153_22": 1,
|
| 1339 |
+
"sub-verse153_23": 2,
|
| 1340 |
+
"sub-verse153_24": 0,
|
| 1341 |
+
"sub-verse400_series1_10": 0,
|
| 1342 |
+
"sub-verse400_series1_11": 0,
|
| 1343 |
+
"sub-verse400_series1_12": 0,
|
| 1344 |
+
"sub-verse400_series1_13": 0,
|
| 1345 |
+
"sub-verse400_series1_14": 3,
|
| 1346 |
+
"sub-verse400_series1_15": 0,
|
| 1347 |
+
"sub-verse400_series1_16": 3,
|
| 1348 |
+
"sub-verse400_series1_17": 0,
|
| 1349 |
+
"sub-verse400_series1_18": 0,
|
| 1350 |
+
"sub-verse400_series1_19": 0,
|
| 1351 |
+
"sub-verse400_series1_20": 2,
|
| 1352 |
+
"sub-verse400_series1_21": 2,
|
| 1353 |
+
"sub-verse400_series1_22": 3,
|
| 1354 |
+
"sub-verse400_series1_23": 0,
|
| 1355 |
+
"sub-verse400_series1_24": 0,
|
| 1356 |
+
"sub-verse404_series0_8": 0,
|
| 1357 |
+
"sub-verse404_series0_9": 0,
|
| 1358 |
+
"sub-verse221_8": 0,
|
| 1359 |
+
"sub-verse221_9": 0,
|
| 1360 |
+
"sub-verse225_8": 0,
|
| 1361 |
+
"sub-verse225_9": 0,
|
| 1362 |
+
"sub-verse230_8": 0,
|
| 1363 |
+
"sub-verse230_9": 0,
|
| 1364 |
+
"sub-verse412_series0_8": 0,
|
| 1365 |
+
"sub-verse412_series0_9": 0,
|
| 1366 |
+
"sub-verse242_8": 0,
|
| 1367 |
+
"sub-verse252_8": 0,
|
| 1368 |
+
"sub-verse252_9": 0,
|
| 1369 |
+
"sub-verse252_10": 0,
|
| 1370 |
+
"sub-verse252_11": 0,
|
| 1371 |
+
"sub-verse252_12": 0,
|
| 1372 |
+
"sub-verse252_13": 0,
|
| 1373 |
+
"sub-verse252_14": 0,
|
| 1374 |
+
"sub-verse252_15": 0,
|
| 1375 |
+
"sub-verse252_16": 0,
|
| 1376 |
+
"sub-verse252_17": 0,
|
| 1377 |
+
"sub-verse252_18": 0,
|
| 1378 |
+
"sub-verse252_19": 0,
|
| 1379 |
+
"sub-verse252_20": 0,
|
| 1380 |
+
"sub-verse252_21": 0,
|
| 1381 |
+
"sub-verse252_22": 0,
|
| 1382 |
+
"sub-verse252_23": 0,
|
| 1383 |
+
"sub-verse404_series1_8": 0,
|
| 1384 |
+
"sub-verse404_series1_9": 0,
|
| 1385 |
+
"sub-verse404_series1_10": 0,
|
| 1386 |
+
"sub-verse404_series1_11": 0,
|
| 1387 |
+
"sub-verse404_series1_12": 0,
|
| 1388 |
+
"sub-verse404_series1_13": 0,
|
| 1389 |
+
"sub-verse404_series1_14": 0,
|
| 1390 |
+
"sub-verse404_series1_15": 0,
|
| 1391 |
+
"sub-verse404_series1_16": 0,
|
| 1392 |
+
"sub-verse404_series1_17": 0,
|
| 1393 |
+
"sub-verse404_series1_18": 0,
|
| 1394 |
+
"sub-verse404_series1_19": 0,
|
| 1395 |
+
"sub-verse404_series1_20": 0,
|
| 1396 |
+
"sub-verse404_series1_21": 0,
|
| 1397 |
+
"sub-verse404_series1_22": 0,
|
| 1398 |
+
"sub-verse404_series1_23": 0,
|
| 1399 |
+
"sub-verse404_series1_24": 0,
|
| 1400 |
+
"sub-verse264_8": 0,
|
| 1401 |
+
"sub-verse264_9": 0,
|
| 1402 |
+
"sub-verse264_10": 0,
|
| 1403 |
+
"sub-verse264_11": 0,
|
| 1404 |
+
"sub-verse264_12": 0,
|
| 1405 |
+
"sub-verse264_13": 0,
|
| 1406 |
+
"sub-verse264_14": 0,
|
| 1407 |
+
"sub-verse264_15": 0,
|
| 1408 |
+
"sub-verse264_16": 0,
|
| 1409 |
+
"sub-verse264_17": 0,
|
| 1410 |
+
"sub-verse264_18": 0,
|
| 1411 |
+
"sub-verse264_19": 0,
|
| 1412 |
+
"sub-verse264_20": 3,
|
| 1413 |
+
"sub-verse264_21": 0,
|
| 1414 |
+
"sub-verse264_22": 0,
|
| 1415 |
+
"sub-verse264_23": 0,
|
| 1416 |
+
"sub-verse264_24": 0,
|
| 1417 |
+
"sub-verse269_9": 0,
|
| 1418 |
+
"sub-verse269_10": 0,
|
| 1419 |
+
"sub-verse269_11": 0,
|
| 1420 |
+
"sub-verse269_12": 0,
|
| 1421 |
+
"sub-verse269_13": 0,
|
| 1422 |
+
"sub-verse269_14": 0,
|
| 1423 |
+
"sub-verse269_15": 0,
|
| 1424 |
+
"sub-verse269_16": 1,
|
| 1425 |
+
"sub-verse269_17": 1,
|
| 1426 |
+
"sub-verse269_18": 0,
|
| 1427 |
+
"sub-verse269_19": 0,
|
| 1428 |
+
"sub-verse269_20": 0,
|
| 1429 |
+
"sub-verse269_21": 0,
|
| 1430 |
+
"sub-verse269_22": 1,
|
| 1431 |
+
"sub-verse269_23": 1,
|
| 1432 |
+
"sub-verse269_24": 0,
|
| 1433 |
+
"sub-verse276_8": 0,
|
| 1434 |
+
"sub-verse276_9": 0,
|
| 1435 |
+
"sub-verse276_10": 0,
|
| 1436 |
+
"sub-verse276_11": 0,
|
| 1437 |
+
"sub-verse276_12": 0,
|
| 1438 |
+
"sub-verse276_13": 0,
|
| 1439 |
+
"sub-verse276_14": 0,
|
| 1440 |
+
"sub-verse276_15": 0,
|
| 1441 |
+
"sub-verse276_16": 0,
|
| 1442 |
+
"sub-verse276_17": 0,
|
| 1443 |
+
"sub-verse276_18": 0,
|
| 1444 |
+
"sub-verse276_19": 1,
|
| 1445 |
+
"sub-verse276_20": 0,
|
| 1446 |
+
"sub-verse276_21": 0,
|
| 1447 |
+
"sub-verse276_22": 0,
|
| 1448 |
+
"sub-verse276_23": 0,
|
| 1449 |
+
"sub-verse276_24": 0,
|
| 1450 |
+
"sub-verse412_series1_8": 0,
|
| 1451 |
+
"sub-verse412_series1_9": 0,
|
| 1452 |
+
"sub-verse412_series1_10": 0,
|
| 1453 |
+
"sub-verse412_series1_11": 0,
|
| 1454 |
+
"sub-verse412_series1_12": 0,
|
| 1455 |
+
"sub-verse412_series1_13": 0,
|
| 1456 |
+
"sub-verse412_series1_14": 0,
|
| 1457 |
+
"sub-verse412_series1_15": 1,
|
| 1458 |
+
"sub-verse412_series1_16": 1,
|
| 1459 |
+
"sub-verse412_series1_17": 0,
|
| 1460 |
+
"sub-verse412_series1_18": 0,
|
| 1461 |
+
"sub-verse412_series1_19": 2,
|
| 1462 |
+
"sub-verse412_series1_20": 1,
|
| 1463 |
+
"sub-verse412_series1_21": 2,
|
| 1464 |
+
"sub-verse412_series1_22": 1,
|
| 1465 |
+
"sub-verse412_series1_23": 0,
|
| 1466 |
+
"sub-verse412_series1_24": 0
|
| 1467 |
+
}
|
| 1468 |
+
}
|
datasets/raw/0007/0007.json
ADDED
|
@@ -0,0 +1,50 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[
|
| 2 |
+
{
|
| 3 |
+
"label": 17,
|
| 4 |
+
"X": 4.949121881359765,
|
| 5 |
+
"Y": 105.01303987337002,
|
| 6 |
+
"Z": 98.28936458882943
|
| 7 |
+
},
|
| 8 |
+
{
|
| 9 |
+
"label": 18,
|
| 10 |
+
"X": 22.963917117592715,
|
| 11 |
+
"Y": 104.28135145836083,
|
| 12 |
+
"Z": 98.88407021248516
|
| 13 |
+
},
|
| 14 |
+
{
|
| 15 |
+
"label": 19,
|
| 16 |
+
"X": 50.349900730980956,
|
| 17 |
+
"Y": 102.38405829798755,
|
| 18 |
+
"Z": 100.23161267033662
|
| 19 |
+
},
|
| 20 |
+
{
|
| 21 |
+
"label": 20,
|
| 22 |
+
"X": 77.50633167353335,
|
| 23 |
+
"Y": 102.35106033415059,
|
| 24 |
+
"Z": 100.20703687789096
|
| 25 |
+
},
|
| 26 |
+
{
|
| 27 |
+
"label": 21,
|
| 28 |
+
"X": 104.47138613177349,
|
| 29 |
+
"Y": 95.11139257996913,
|
| 30 |
+
"Z": 101.01669622089676
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"label": 22,
|
| 34 |
+
"X": 132.90240595760932,
|
| 35 |
+
"Y": 87.06807332442239,
|
| 36 |
+
"Z": 99.48401756730952
|
| 37 |
+
},
|
| 38 |
+
{
|
| 39 |
+
"label": 23,
|
| 40 |
+
"X": 161.00800884787003,
|
| 41 |
+
"Y": 80.13662713092559,
|
| 42 |
+
"Z": 96.85088288013425
|
| 43 |
+
},
|
| 44 |
+
{
|
| 45 |
+
"label": 24,
|
| 46 |
+
"X": 188.4172078227643,
|
| 47 |
+
"Y": 75.91908069343272,
|
| 48 |
+
"Z": 93.20695691456322
|
| 49 |
+
}
|
| 50 |
+
]
|
datasets/raw/0007/0007.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a357509a09b696b94ce923775e3638204df5097cc077cc3c4b95686e3bb34a39
|
| 3 |
+
size 13843578
|
datasets/raw/0007/0007_msk.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c319b8334a76518466e3aa914860f7e30becd157c6f7e544198ed097f2a0b97a
|
| 3 |
+
size 195059
|
datasets/straightened/CT/0007_18.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4f5b6d714c03a37603a9b04b04a4859db4ae62cb3845346c25ff1fb26f3bad11
|
| 3 |
+
size 9363428
|
datasets/straightened/CT/0007_19.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:81dd9cc077b7145db893634323652f9f82843924c15f0f7726a86cbbaf41ed2b
|
| 3 |
+
size 10986869
|
datasets/straightened/CT/0007_20.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:759bd6f239efeb4e054408b2be3098ff797168e561ad588d2848cd4a6f06295f
|
| 3 |
+
size 12616657
|
datasets/straightened/CT/0007_21.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:9e25b4a26709f67bf9a980e46437b6686d88f990d78e31a63af4f7aaebc9a4e3
|
| 3 |
+
size 13103420
|
datasets/straightened/CT/0007_22.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cd84db5a2a0e6e8808a1e5750dee51650b0e217957cde314f0e3b428a2712ad1
|
| 3 |
+
size 12793166
|
datasets/straightened/CT/0007_23.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6bb2b54b6ad32536e189f3209fbb041d95f1eb7d259c71ed8caa2047f66e92ea
|
| 3 |
+
size 11081586
|
datasets/straightened/label/0007_18.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:beb2e06f38bbf1cc695b7cde3b59f40fa67215eca2bfd7b49a384de125f40700
|
| 3 |
+
size 222229
|
datasets/straightened/label/0007_19.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:316b886125a6f9323c08f21ab91cc93da414ea1a6ae2225eb30479921e64ab34
|
| 3 |
+
size 238415
|
datasets/straightened/label/0007_20.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:24e7bab70bdb78cb2f7f6eaf43edd91f44ae0ed342c9c07988661a1970508d49
|
| 3 |
+
size 252946
|
datasets/straightened/label/0007_21.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f2002efa3e5482c8ca7803cd92f9506bbaed086006f3409c7888fb3626901dc8
|
| 3 |
+
size 253722
|
datasets/straightened/label/0007_22.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:32b1c5705bc3d2c5aefa28d17d597a454093a67d44c0152ed8b75476b895526e
|
| 3 |
+
size 249093
|
datasets/straightened/label/0007_23.nii.gz
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0bd43be84dcdd4482f9b0e3670cb287619fab7fa9a1983ddf2f84696a16dc1f5
|
| 3 |
+
size 234749
|
eval_3d_sagittal_twostage.py
ADDED
|
@@ -0,0 +1,263 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#先生成目标椎体上下的椎体,再对目标椎体做生成
|
| 2 |
+
|
| 3 |
+
import torch
|
| 4 |
+
import numpy as np
|
| 5 |
+
import nibabel as nib
|
| 6 |
+
import os
|
| 7 |
+
from options.test_options import TestOptions
|
| 8 |
+
from models import create_model
|
| 9 |
+
import torchvision.transforms as transforms
|
| 10 |
+
from PIL import Image
|
| 11 |
+
from models.inpaint_networks import Generator
|
| 12 |
+
import torch.nn.functional as F
|
| 13 |
+
import math
|
| 14 |
+
from scipy.ndimage import label
|
| 15 |
+
|
| 16 |
+
def remove_small_connected_components(input_array, min_size):
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
# 识别连通域
|
| 20 |
+
structure = np.ones((3, 3), dtype=np.int32) # 定义连通性结构
|
| 21 |
+
labeled, ncomponents = label(input_array, structure)
|
| 22 |
+
|
| 23 |
+
# 遍历所有连通域,如果连通域大小小于阈值,则去除
|
| 24 |
+
for i in range(1, ncomponents + 1):
|
| 25 |
+
if np.sum(labeled == i) < min_size:
|
| 26 |
+
input_array[labeled == i] = 0
|
| 27 |
+
|
| 28 |
+
# 如果输入是张量,则转换回张量
|
| 29 |
+
|
| 30 |
+
return input_array
|
| 31 |
+
|
| 32 |
+
def load_model(model_path, netG_params, device):
|
| 33 |
+
model = Generator(netG_params, True)
|
| 34 |
+
if os.path.exists(model_path):
|
| 35 |
+
model.load_state_dict(torch.load(model_path, map_location=device))
|
| 36 |
+
model.eval()
|
| 37 |
+
model.to(device)
|
| 38 |
+
return model
|
| 39 |
+
|
| 40 |
+
def numpy_to_pil(img_np):
|
| 41 |
+
if img_np.dtype != np.uint8:
|
| 42 |
+
raise ValueError("NumPy array should have uint8 data type.")
|
| 43 |
+
img_pil = Image.fromarray(img_np)
|
| 44 |
+
return img_pil
|
| 45 |
+
|
| 46 |
+
def run_model(model,CAM_data,label_data,ct_data,vert_id,index_ratio,A_transform,mask_transform,device,maxheight=40):
|
| 47 |
+
vert_label_slice = np.zeros_like(label_data)
|
| 48 |
+
vert_label_slice[label_data==vert_id]=1
|
| 49 |
+
|
| 50 |
+
vert_label_slice = remove_small_connected_components(vert_label_slice,50)
|
| 51 |
+
coords = np.argwhere(vert_label_slice)
|
| 52 |
+
if coords.size==0:
|
| 53 |
+
return None
|
| 54 |
+
x1, x2 = min(coords[:, 0]), max(coords[:, 0])
|
| 55 |
+
width,length = vert_label_slice.shape
|
| 56 |
+
height = x2-x1
|
| 57 |
+
if height>maxheight:
|
| 58 |
+
x_mean = int(np.mean(coords[:, 0]))
|
| 59 |
+
x1 = x_mean-20
|
| 60 |
+
x2 = x1+40
|
| 61 |
+
|
| 62 |
+
mask_x = (x1+x2)//2
|
| 63 |
+
h2 = maxheight
|
| 64 |
+
if mask_x<=h2//2:
|
| 65 |
+
min_x = 0
|
| 66 |
+
max_x = min_x + h2
|
| 67 |
+
elif width-mask_x<=h2/2:
|
| 68 |
+
max_x = width
|
| 69 |
+
min_x = max_x -h2
|
| 70 |
+
else:
|
| 71 |
+
min_x = mask_x-h2//2
|
| 72 |
+
max_x = min_x + h2
|
| 73 |
+
|
| 74 |
+
mask_slice = np.zeros_like(vert_label_slice).astype(np.uint8)
|
| 75 |
+
mask_slice[min_x:max_x+1] = 255
|
| 76 |
+
ct_data_slice = np.zeros_like(mask_slice).astype(np.uint8)
|
| 77 |
+
ct_data_slice[:min_x,:] = ct_data[(x1-min_x):x1,:]
|
| 78 |
+
ct_data_slice[max_x:,:] = ct_data[x2:x2+(width-max_x),:]
|
| 79 |
+
|
| 80 |
+
CAM_slice = np.zeros_like(mask_slice).astype(np.uint8)
|
| 81 |
+
CAM_slice[:min_x,:] = CAM_data[(x1-min_x):x1,:]
|
| 82 |
+
CAM_slice[max_x:,:] = CAM_data[x2:x2+(width-max_x),:]
|
| 83 |
+
|
| 84 |
+
ct_batch = numpy_to_pil(ct_data_slice)
|
| 85 |
+
ct_batch = A_transform(ct_batch)
|
| 86 |
+
|
| 87 |
+
ori_ct = numpy_to_pil(ct_data.astype(np.uint8))
|
| 88 |
+
ori_ct = A_transform(ori_ct)
|
| 89 |
+
|
| 90 |
+
mask_batch = numpy_to_pil(mask_slice)
|
| 91 |
+
mask_batch = mask_transform(mask_batch)
|
| 92 |
+
|
| 93 |
+
CAM = numpy_to_pil(CAM_slice)
|
| 94 |
+
CAM = mask_transform(CAM)
|
| 95 |
+
|
| 96 |
+
ct_batch = ct_batch.unsqueeze(0).to(device)
|
| 97 |
+
mask_batch = mask_batch.unsqueeze(0).to(device)
|
| 98 |
+
CAM = CAM.unsqueeze(0).to(device)
|
| 99 |
+
|
| 100 |
+
with torch.no_grad():
|
| 101 |
+
_, fake_B_mask_sigmoid, _, fake_B_raw, _,_,pred_h = model(ct_batch, mask_batch, 1-CAM,index_ratio)
|
| 102 |
+
#print(pred_h)
|
| 103 |
+
pred_h = math.ceil(pred_h[0]*maxheight)
|
| 104 |
+
|
| 105 |
+
fake_B_mask_raw = torch.where(fake_B_mask_sigmoid > 0.5, torch.ones_like(fake_B_mask_sigmoid), torch.zeros_like(fake_B_mask_sigmoid))
|
| 106 |
+
#fake_B_mask_raw = fake_B_mask_raw.squeeze().cpu().numpy()*int(vert_id)
|
| 107 |
+
|
| 108 |
+
if pred_h<height:
|
| 109 |
+
pred_h = height
|
| 110 |
+
height_diff = pred_h-height
|
| 111 |
+
x_upper = x1-height_diff//2
|
| 112 |
+
x_bottom = x_upper+pred_h
|
| 113 |
+
single_image = torch.zeros_like(fake_B_raw)
|
| 114 |
+
single_image[:,:,x_upper:x_bottom,:] = fake_B_raw[:,:,x_upper:x_bottom,:]
|
| 115 |
+
ct_upper = torch.zeros_like(single_image)
|
| 116 |
+
ct_upper[0,:,:x_upper,:] = ori_ct[:, height_diff//2:x1, :]
|
| 117 |
+
ct_bottom = torch.zeros_like(single_image)
|
| 118 |
+
ct_bottom[0,:,x_bottom:,:] = ori_ct[:, x2:x2+256-x_bottom, :]
|
| 119 |
+
interpolated_image = single_image+ct_upper+ct_bottom
|
| 120 |
+
fake_B = interpolated_image.squeeze().cpu().numpy()
|
| 121 |
+
fake_B = (fake_B+1)*127.5
|
| 122 |
+
|
| 123 |
+
mid_seg = np.zeros_like(fake_B_mask_raw.squeeze().cpu().numpy())
|
| 124 |
+
mid_seg[x_upper:x_bottom,:] = fake_B_mask_raw[:,:,x_upper:x_bottom,:].squeeze().cpu().numpy()*vert_id
|
| 125 |
+
seg_upper = np.zeros_like(mid_seg)
|
| 126 |
+
seg_upper[:x_upper,:] = label_data[height_diff//2:x1, :]
|
| 127 |
+
seg_bottom = np.zeros_like(mid_seg)
|
| 128 |
+
seg_bottom[x_bottom:,:] = label_data[x2:x2+256-x_bottom, :]
|
| 129 |
+
interpolated_seg = mid_seg+seg_upper+seg_bottom
|
| 130 |
+
fake_B_mask_raw = interpolated_seg
|
| 131 |
+
|
| 132 |
+
|
| 133 |
+
return fake_B_mask_raw,fake_B,height
|
| 134 |
+
|
| 135 |
+
|
| 136 |
+
def process_nii_files(folder_path,CAM_folder, model, output_folder, device):
|
| 137 |
+
A_transform = transforms.Compose([
|
| 138 |
+
transforms.Grayscale(1),
|
| 139 |
+
transforms.ToTensor(),
|
| 140 |
+
transforms.Normalize((0.5,), (0.5,))
|
| 141 |
+
])
|
| 142 |
+
|
| 143 |
+
mask_transform = transforms.Compose([
|
| 144 |
+
transforms.ToTensor()
|
| 145 |
+
])
|
| 146 |
+
|
| 147 |
+
if not os.path.exists(os.path.join(output_folder, 'CT')):
|
| 148 |
+
os.makedirs(os.path.join(output_folder, 'CT'))
|
| 149 |
+
if not os.path.exists(os.path.join(output_folder, 'label')):
|
| 150 |
+
os.makedirs(os.path.join(output_folder, 'label'))
|
| 151 |
+
|
| 152 |
+
count = 0
|
| 153 |
+
for file_name in os.listdir(folder_path):
|
| 154 |
+
#if file_name!="sub-verse013_22.nii.gz":
|
| 155 |
+
# continue
|
| 156 |
+
if file_name.endswith('.nii.gz'):
|
| 157 |
+
if os.path.exists(os.path.join(output_folder, 'CT_fake', file_name)):
|
| 158 |
+
continue
|
| 159 |
+
#if file_name!="sub-verse004_20.nii.gz":
|
| 160 |
+
# continue
|
| 161 |
+
file_path = os.path.join(folder_path, file_name)
|
| 162 |
+
label_path = file_path.replace('CT', 'label')
|
| 163 |
+
ct_nii = nib.load(file_path)
|
| 164 |
+
ct_data = ct_nii.get_fdata()
|
| 165 |
+
label_nii = nib.load(label_path)
|
| 166 |
+
label_data = label_nii.get_fdata()
|
| 167 |
+
patient_id, vert_id = file_name[:-7].rsplit('_', 1)
|
| 168 |
+
vert_id = int(vert_id)
|
| 169 |
+
|
| 170 |
+
CAM_path_0 = os.path.join(CAM_folder, file_name[:-7]+'_0.nii.gz')
|
| 171 |
+
CAM_path_1 = os.path.join(CAM_folder, file_name[:-7]+'_1.nii.gz')
|
| 172 |
+
CAM_path_2 = os.path.join(CAM_folder, file_name[:-7]+'.nii.gz')
|
| 173 |
+
if os.path.exists(CAM_path_0):
|
| 174 |
+
CAM_path = CAM_path_0
|
| 175 |
+
elif os.path.exists(CAM_path_1):
|
| 176 |
+
CAM_path = CAM_path_1
|
| 177 |
+
else:
|
| 178 |
+
CAM_path = CAM_path_2
|
| 179 |
+
|
| 180 |
+
#print(CAM_path)
|
| 181 |
+
CAM_data = nib.load(CAM_path).get_fdata() * 255
|
| 182 |
+
|
| 183 |
+
vert_label = np.zeros_like(label_data)
|
| 184 |
+
vert_label[label_data==vert_id]=1
|
| 185 |
+
|
| 186 |
+
loc = np.where(vert_label)
|
| 187 |
+
|
| 188 |
+
z0 = min(loc[2])
|
| 189 |
+
z1 = max(loc[2])
|
| 190 |
+
range_length = z1 - z0 + 1
|
| 191 |
+
new_range_length = int(range_length * 4 / 5)
|
| 192 |
+
new_z0 = z0 + (range_length - new_range_length) // 2
|
| 193 |
+
new_z1 = new_z0 + new_range_length - 1
|
| 194 |
+
|
| 195 |
+
output_ct_data = np.zeros_like(ct_data)
|
| 196 |
+
output_seg_data = np.zeros_like(ct_data)
|
| 197 |
+
center_index = (new_z0 + new_z1) // 2
|
| 198 |
+
|
| 199 |
+
maxheight = 40
|
| 200 |
+
|
| 201 |
+
for z in range(new_z0, new_z1 + 1):
|
| 202 |
+
index_ratio = abs(z-center_index)/range_length*2
|
| 203 |
+
index_ratio = torch.tensor([index_ratio])
|
| 204 |
+
if int(vert_id)>8 and np.sum(label_data[:, :, z]==int(vert_id)-1)>200:
|
| 205 |
+
#print("upper exists and sum=",np.sum(label_data[:, :, z]==int(vert_id)-1))
|
| 206 |
+
vert_id_upper = int(vert_id)-1
|
| 207 |
+
#print("upper exists")
|
| 208 |
+
fake_B_mask_upper,fake_B_ct_upper,_ = run_model(model,CAM_data[:, :, z],label_data[:, :, z],ct_data[:, :, z],vert_id_upper,index_ratio,\
|
| 209 |
+
A_transform,mask_transform,device,maxheight)
|
| 210 |
+
else:
|
| 211 |
+
fake_B_mask_upper,fake_B_ct_upper = label_data[:, :, z],ct_data[:, :, z]
|
| 212 |
+
#print("upper dont exists and sum=",np.sum(label_data[:, :, z]==int(vert_id)-1))
|
| 213 |
+
if int(vert_id)<24 and np.sum(label_data[:, :, z]==int(vert_id)+1)>200:
|
| 214 |
+
#print("bottom exists and sum=",np.sum(label_data[:, :, z]==int(vert_id)+1))
|
| 215 |
+
vert_id_bottom = int(vert_id)+1
|
| 216 |
+
#print("bottom exists")
|
| 217 |
+
fake_B_mask_bottom,fake_B_ct_bottom,_ = run_model(model,CAM_data[:, :, z],fake_B_mask_upper,fake_B_ct_upper,vert_id_bottom,index_ratio,\
|
| 218 |
+
A_transform,mask_transform,device,maxheight)
|
| 219 |
+
else:
|
| 220 |
+
fake_B_mask_bottom,fake_B_ct_bottom = fake_B_mask_upper,fake_B_ct_upper
|
| 221 |
+
#print("bottom dont exists and sum=",np.sum(label_data[:, :, z]==int(vert_id)+1))
|
| 222 |
+
|
| 223 |
+
|
| 224 |
+
output = run_model(model,CAM_data[:, :, z],fake_B_mask_bottom,fake_B_ct_bottom,int(vert_id),index_ratio,\
|
| 225 |
+
A_transform,mask_transform,device,maxheight)
|
| 226 |
+
if output==None:
|
| 227 |
+
continue
|
| 228 |
+
else:
|
| 229 |
+
fake_B_mask_raw,fake_B,height = output
|
| 230 |
+
if height>maxheight:
|
| 231 |
+
print("Height exceeds in %s, in slice %d"%(file_name,z))
|
| 232 |
+
|
| 233 |
+
output_seg_data[:, :, z] = fake_B_mask_raw
|
| 234 |
+
output_ct_data[:, :, z] = fake_B
|
| 235 |
+
|
| 236 |
+
new_ct_nii = nib.Nifti1Image(output_ct_data, ct_nii.affine)
|
| 237 |
+
nib.save(new_ct_nii, os.path.join(output_folder, 'CT_fake', file_name))
|
| 238 |
+
new_label_nii = nib.Nifti1Image(output_seg_data, ct_nii.affine)
|
| 239 |
+
nib.save(new_label_nii, os.path.join(output_folder, 'label_fake', file_name))
|
| 240 |
+
print(f"Now {file_name} has been generateed in {output_folder}")
|
| 241 |
+
count+=1
|
| 242 |
+
|
| 243 |
+
|
| 244 |
+
def main():
|
| 245 |
+
model_path = '/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/checkpoints/0421_adaptive_sagittal/latest_net_G.pth'
|
| 246 |
+
netG_params = {'input_dim': 1, 'ngf': 16}
|
| 247 |
+
#folder_path = '/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/datasets/straighten/revised/CT'
|
| 248 |
+
#CAM_folder = '/home/zhangqi/Project/VertebralFractureGrading/heatmap/straighten_sagittal/binaryclass_1'
|
| 249 |
+
#output_folder = '/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/output_3d/sagittal/fine'
|
| 250 |
+
folder_path = '/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/datasets/local/straighten/CT'
|
| 251 |
+
CAM_folder = '/home/zhangqi/Project/VertebralFractureGrading/heatmap/local_sagittal_0508/binaryclass_1'
|
| 252 |
+
output_folder = '/home/zhangqi/Project/pytorch-CycleGAN-and-pix2pix-master/output_3d/local_dataset/sagittal/fine'
|
| 253 |
+
if not os.path.exists(output_folder+'/CT_fake'):
|
| 254 |
+
os.makedirs(output_folder+'/CT_fake')
|
| 255 |
+
if not os.path.exists(output_folder+'/label_fake'):
|
| 256 |
+
os.makedirs(output_folder+'/label_fake')
|
| 257 |
+
device = 'cuda:0'
|
| 258 |
+
|
| 259 |
+
model = load_model(model_path, netG_params, device)
|
| 260 |
+
process_nii_files(folder_path,CAM_folder, model, output_folder, device)
|
| 261 |
+
|
| 262 |
+
if __name__ == "__main__":
|
| 263 |
+
main()
|
evaluation/RHLV_quantification.py
ADDED
|
@@ -0,0 +1,212 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import json
|
| 2 |
+
import numpy as np
|
| 3 |
+
import os
|
| 4 |
+
import nibabel as nib
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import cv2
|
| 7 |
+
import pandas as pd
|
| 8 |
+
from sklearn.model_selection import ParameterGrid
|
| 9 |
+
|
| 10 |
+
def rotate_image_to_horizontal(binary_image):
|
| 11 |
+
"""
|
| 12 |
+
Rotates the image to make the major axis of the object horizontal.
|
| 13 |
+
"""
|
| 14 |
+
# 寻找轮廓
|
| 15 |
+
binary_image = binary_image.astype(np.uint8)
|
| 16 |
+
contours, _ = cv2.findContours(binary_image, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
| 17 |
+
|
| 18 |
+
# 假设最大的轮廓是椎体
|
| 19 |
+
contour = max(contours, key=cv2.contourArea)
|
| 20 |
+
|
| 21 |
+
# 获取轮廓的最小外接矩形
|
| 22 |
+
rect = cv2.minAreaRect(contour)
|
| 23 |
+
box = cv2.boxPoints(rect)
|
| 24 |
+
box = np.int0(box)
|
| 25 |
+
|
| 26 |
+
# 计算旋转角度
|
| 27 |
+
angle = rect[2]
|
| 28 |
+
if angle < -45:
|
| 29 |
+
angle += 90
|
| 30 |
+
if angle > 45:
|
| 31 |
+
angle-=90
|
| 32 |
+
|
| 33 |
+
# 旋转图像
|
| 34 |
+
(h, w) = binary_image.shape[:2]
|
| 35 |
+
center = (w // 2, h // 2)
|
| 36 |
+
M = cv2.getRotationMatrix2D(center, angle, 1.0)
|
| 37 |
+
rotated_image = cv2.warpAffine(binary_image, M, (w, h), flags=cv2.INTER_CUBIC, borderMode=cv2.BORDER_CONSTANT)
|
| 38 |
+
|
| 39 |
+
return rotated_image
|
| 40 |
+
|
| 41 |
+
def calculate_heights(segmentation_fake,segmentation_label,height_threshold):
|
| 42 |
+
all_heights_fake = []
|
| 43 |
+
all_heights_label = []
|
| 44 |
+
pre_heights_fake = []
|
| 45 |
+
pre_heights_label = []
|
| 46 |
+
mid_heights_fake = []
|
| 47 |
+
mid_heights_label = []
|
| 48 |
+
post_heights_fake = []
|
| 49 |
+
post_heights_label = []
|
| 50 |
+
# 遍历z轴上的每个层
|
| 51 |
+
for z in range(segmentation_label.shape[2]):
|
| 52 |
+
if np.any(segmentation_label[:, :, z]) and np.any(segmentation_fake[:, :, z]):
|
| 53 |
+
segmentation_label_slice = segmentation_label[:, :, z]
|
| 54 |
+
segmentation_fake_slice = segmentation_fake[:, :, z]
|
| 55 |
+
|
| 56 |
+
loc = np.where(segmentation_fake_slice)[1]
|
| 57 |
+
y_min = int(np.min(loc))
|
| 58 |
+
y_max = int(np.max(loc))
|
| 59 |
+
y_range = y_max-y_min
|
| 60 |
+
one_third_y = int(y_min + y_range/3)
|
| 61 |
+
two_third_y = int(y_min + 2*y_range/3)
|
| 62 |
+
center_height_fake = np.count_nonzero(segmentation_fake_slice[:, int(np.mean(loc))])
|
| 63 |
+
all_height_fake = np.count_nonzero(segmentation_fake_slice, axis=0)
|
| 64 |
+
pre_height_fake = np.count_nonzero(segmentation_fake_slice[:,:one_third_y], axis=0)
|
| 65 |
+
mid_height_fake = np.count_nonzero(segmentation_fake_slice[:,one_third_y:two_third_y], axis=0)
|
| 66 |
+
post_height_fake = np.count_nonzero(segmentation_fake_slice[:, two_third_y:], axis=0)
|
| 67 |
+
|
| 68 |
+
loc = np.where(segmentation_label[:, :, z])[1]
|
| 69 |
+
center_height_label = np.count_nonzero(segmentation_label_slice[:, int(np.mean(loc))])
|
| 70 |
+
all_height_label = np.count_nonzero(segmentation_label_slice, axis=0)
|
| 71 |
+
pre_height_label = np.count_nonzero(segmentation_label_slice[:, :one_third_y], axis=0)
|
| 72 |
+
mid_height_label = np.count_nonzero(segmentation_label_slice[:, one_third_y:two_third_y], axis=0)
|
| 73 |
+
post_height_label = np.count_nonzero(segmentation_label_slice[:, two_third_y:], axis=0)
|
| 74 |
+
|
| 75 |
+
|
| 76 |
+
all_scale_ratio = 1
|
| 77 |
+
pre_scale_ratio = 1
|
| 78 |
+
mid_scale_ratio = 1
|
| 79 |
+
post_scale_ratio = 1
|
| 80 |
+
if all_height_label.size > 0 and all_height_fake.size > 0:
|
| 81 |
+
if all_height_label.max()>all_height_fake.max():
|
| 82 |
+
all_scale_ratio = all_height_label.max()/(all_height_fake.max()+1e-6)
|
| 83 |
+
if pre_height_label.size > 0 and pre_height_fake.size > 0:
|
| 84 |
+
if pre_height_label.max()>pre_height_fake.max():
|
| 85 |
+
pre_scale_ratio = pre_height_label.max()/(pre_height_fake.max()+1e-6)
|
| 86 |
+
if mid_height_label.size > 0 and mid_height_fake.size > 0:
|
| 87 |
+
if mid_height_label.max()>mid_height_fake.max():
|
| 88 |
+
mid_scale_ratio = mid_height_label.max()/(mid_height_fake.max()+1e-6)
|
| 89 |
+
if post_height_label.size > 0 and post_height_fake.size > 0:
|
| 90 |
+
if post_height_label.max()>post_height_fake.max():
|
| 91 |
+
post_scale_ratio = post_height_label.max()/(post_height_fake.max()+1e-6)
|
| 92 |
+
|
| 93 |
+
all_height_fake = all_height_fake*all_scale_ratio
|
| 94 |
+
center_height_fake = center_height_fake*all_scale_ratio
|
| 95 |
+
pre_height_fake = pre_height_fake*pre_scale_ratio
|
| 96 |
+
mid_height_fake = mid_height_fake*mid_scale_ratio
|
| 97 |
+
post_height_fake = post_height_fake*post_scale_ratio
|
| 98 |
+
|
| 99 |
+
all_heights_fake.extend(all_height_fake[all_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 100 |
+
all_heights_label.extend(all_height_label[all_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 101 |
+
pre_heights_fake.extend(pre_height_fake[pre_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 102 |
+
pre_heights_label.extend(pre_height_label[pre_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 103 |
+
mid_heights_fake.extend(mid_height_fake[mid_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 104 |
+
mid_heights_label.extend(mid_height_label[mid_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 105 |
+
post_heights_fake.extend(post_height_fake[post_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 106 |
+
post_heights_label.extend(post_height_label[post_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 107 |
+
|
| 108 |
+
# 将heights转换为numpy数组以便使用numpy的功能
|
| 109 |
+
all_heights_fake = np.array(all_heights_fake)
|
| 110 |
+
all_heights_label = np.array(all_heights_label)
|
| 111 |
+
pre_heights_fake = np.array(pre_heights_fake)
|
| 112 |
+
pre_heights_label = np.array(pre_heights_label)
|
| 113 |
+
mid_heights_fake = np.array(mid_heights_fake)
|
| 114 |
+
mid_heights_label = np.array(mid_heights_label)
|
| 115 |
+
post_heights_fake = np.array(post_heights_fake)
|
| 116 |
+
post_heights_label = np.array(post_heights_label)
|
| 117 |
+
|
| 118 |
+
return all_heights_fake, all_heights_label,pre_heights_fake, pre_heights_label,mid_heights_fake, mid_heights_label,post_heights_fake, post_heights_label
|
| 119 |
+
|
| 120 |
+
|
| 121 |
+
def calculate_rhlv(segmentation_fake, segmentation_label, center_z, length,vertebra,height_threshold):
|
| 122 |
+
"""
|
| 123 |
+
Calculate the Relative Height Loss Value (RHLV) between fake and label segmentations.
|
| 124 |
+
"""
|
| 125 |
+
seg_fake_filtered = segmentation_fake[:, :, center_z-length:center_z+length]
|
| 126 |
+
seg_label_filtered = segmentation_label[:, :, center_z-length:center_z+length]
|
| 127 |
+
|
| 128 |
+
all_heights_fake, all_heights_label,pre_heights_fake, pre_heights_label,mid_heights_fake, mid_heights_label,post_heights_fake, post_heights_label\
|
| 129 |
+
= calculate_heights(seg_fake_filtered, seg_label_filtered,height_threshold)
|
| 130 |
+
all_height_fake = np.mean(all_heights_fake) if all_heights_fake.size > 0 else 0
|
| 131 |
+
all_height_label = np.mean(all_heights_label) if all_heights_label.size > 0 else 0
|
| 132 |
+
pre_height_fake = np.mean(pre_heights_fake) if pre_heights_fake.size > 0 else 0
|
| 133 |
+
pre_height_label = np.mean(pre_heights_label) if pre_heights_label.size > 0 else 0
|
| 134 |
+
mid_height_fake = np.mean(mid_heights_fake) if mid_heights_fake.size > 0 else 0
|
| 135 |
+
mid_height_label = np.mean(mid_heights_label) if mid_heights_label.size > 0 else 0
|
| 136 |
+
post_height_fake = np.mean(post_heights_fake) if post_heights_fake.size > 0 else 0
|
| 137 |
+
post_height_label = np.mean(post_heights_label) if post_heights_label.size > 0 else 0
|
| 138 |
+
|
| 139 |
+
all_rhlv = (all_height_fake - all_height_label) / (all_height_fake +1e-6)
|
| 140 |
+
pre_rhlv = (pre_height_fake - pre_height_label) / (pre_height_fake +1e-6)
|
| 141 |
+
mid_rhlv = (mid_height_fake - mid_height_label) / (mid_height_fake +1e-6)
|
| 142 |
+
post_rhlv = (post_height_fake - post_height_label) / (post_height_fake +1e-6)
|
| 143 |
+
min_height = min(pre_height_label,mid_height_label,post_height_label)
|
| 144 |
+
max_height = max(pre_height_label,mid_height_label,post_height_label)
|
| 145 |
+
relative_height_label = min_height/(max_height+1e-6)
|
| 146 |
+
|
| 147 |
+
return all_rhlv,pre_rhlv,mid_rhlv,post_rhlv,relative_height_label
|
| 148 |
+
|
| 149 |
+
def process_datasets_to_excel(dataset_info, label_folder, fake_folder, output_file,length_divisor=5, height_threshold=0.64):
|
| 150 |
+
results = []
|
| 151 |
+
for dataset_type, data in dataset_info.items():
|
| 152 |
+
for vertebra, label in data.items():
|
| 153 |
+
label_path = os.path.join(label_folder, vertebra + '.nii.gz')
|
| 154 |
+
fake_path = os.path.join(fake_folder, vertebra + '.nii.gz')
|
| 155 |
+
|
| 156 |
+
if not os.path.exists(label_path) or not os.path.exists(fake_path):
|
| 157 |
+
continue
|
| 158 |
+
|
| 159 |
+
segmentation_label_temp = nib.load(label_path).get_fdata()
|
| 160 |
+
segmentation_label = np.zeros_like(segmentation_label_temp)
|
| 161 |
+
|
| 162 |
+
segmentation_fake_temp = nib.load(fake_path).get_fdata()
|
| 163 |
+
segmentation_fake = np.zeros_like(segmentation_fake_temp)
|
| 164 |
+
|
| 165 |
+
label_index = int(vertebra.split('_')[-1])
|
| 166 |
+
segmentation_label[segmentation_label_temp == label_index] = 1
|
| 167 |
+
segmentation_fake[segmentation_fake_temp == label_index] = 1
|
| 168 |
+
|
| 169 |
+
loc = np.where(segmentation_label)[2]
|
| 170 |
+
if loc.size == 0:
|
| 171 |
+
continue # Skip if no label index found
|
| 172 |
+
|
| 173 |
+
min_z = np.min(loc)
|
| 174 |
+
max_z = np.max(loc)
|
| 175 |
+
center_z = int(np.mean(loc))
|
| 176 |
+
length = (max_z - min_z) // length_divisor # Divisor adjusted based on your setup
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
all_rhlv, pre_rhlv, mid_rhlv, post_rhlv, relative_height_label = calculate_rhlv(
|
| 180 |
+
segmentation_fake, segmentation_label, center_z, length, vertebra,height_threshold
|
| 181 |
+
)
|
| 182 |
+
results.append({
|
| 183 |
+
"Vertebra": vertebra,
|
| 184 |
+
"Label": label,
|
| 185 |
+
"Dataset": dataset_type,
|
| 186 |
+
"All RHLV": all_rhlv,
|
| 187 |
+
"Pre RHLV": pre_rhlv,
|
| 188 |
+
"Mid RHLV": mid_rhlv,
|
| 189 |
+
"Post RHLV": post_rhlv,
|
| 190 |
+
"Relative Height Label": relative_height_label
|
| 191 |
+
})
|
| 192 |
+
|
| 193 |
+
# Create a DataFrame from results and save to Excel
|
| 194 |
+
df = pd.DataFrame(results)
|
| 195 |
+
df.to_excel(output_file, index=False)
|
| 196 |
+
|
| 197 |
+
def main():
|
| 198 |
+
with open('vertebra_data_local.json', 'r') as file:
|
| 199 |
+
json_data = json.load(file)
|
| 200 |
+
|
| 201 |
+
label_folder = '/dssg/home/acct-milesun/zhangqi/Dataset/HealthiVert_straighten/label'
|
| 202 |
+
output_folder = '/dssg/home/acct-milesun/zhangqi/Project/HealthiVert-GAN_eval/output'
|
| 203 |
+
result_folder = '/dssg/home/acct-milesun/zhangqi/Project/HealthiVert-GAN_eval/evaluation/RHLV_quantification'
|
| 204 |
+
for root, dirs, files in os.walk(output_folder):
|
| 205 |
+
for dir in dirs:
|
| 206 |
+
exp_folder = os.path.join(root,dir)
|
| 207 |
+
fake_folder = os.path.join(exp_folder,'label_fake')
|
| 208 |
+
result_file = os.path.join(result_folder,dir+'.xlsx')
|
| 209 |
+
process_datasets_to_excel(json_data, label_folder, fake_folder, result_file, length_divisor=5, height_threshold=0.7)
|
| 210 |
+
|
| 211 |
+
if __name__ == "__main__":
|
| 212 |
+
main()
|
evaluation/RHLV_quantification_coronal.py
ADDED
|
@@ -0,0 +1,218 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import json
|
| 2 |
+
import numpy as np
|
| 3 |
+
import os
|
| 4 |
+
import nibabel as nib
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import cv2
|
| 7 |
+
import pandas as pd
|
| 8 |
+
from sklearn.model_selection import ParameterGrid
|
| 9 |
+
|
| 10 |
+
def rotate_image_to_horizontal(binary_image):
|
| 11 |
+
"""
|
| 12 |
+
Rotates the image to make the major axis of the object horizontal.
|
| 13 |
+
"""
|
| 14 |
+
# 寻找轮廓
|
| 15 |
+
binary_image = binary_image.astype(np.uint8)
|
| 16 |
+
contours, _ = cv2.findContours(binary_image, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
| 17 |
+
|
| 18 |
+
# 假设最大的轮廓是椎体
|
| 19 |
+
contour = max(contours, key=cv2.contourArea)
|
| 20 |
+
|
| 21 |
+
# 获取轮廓的最小外接矩形
|
| 22 |
+
rect = cv2.minAreaRect(contour)
|
| 23 |
+
box = cv2.boxPoints(rect)
|
| 24 |
+
box = np.int0(box)
|
| 25 |
+
|
| 26 |
+
# 计算旋转角度
|
| 27 |
+
angle = rect[2]
|
| 28 |
+
if angle < -45:
|
| 29 |
+
angle += 90
|
| 30 |
+
if angle > 45:
|
| 31 |
+
angle-=90
|
| 32 |
+
|
| 33 |
+
# 旋转图像
|
| 34 |
+
(h, w) = binary_image.shape[:2]
|
| 35 |
+
center = (w // 2, h // 2)
|
| 36 |
+
M = cv2.getRotationMatrix2D(center, angle, 1.0)
|
| 37 |
+
rotated_image = cv2.warpAffine(binary_image, M, (w, h), flags=cv2.INTER_CUBIC, borderMode=cv2.BORDER_CONSTANT)
|
| 38 |
+
|
| 39 |
+
return rotated_image
|
| 40 |
+
|
| 41 |
+
def calculate_heights(segmentation_fake,segmentation_label,height_threshold):
|
| 42 |
+
all_heights_fake = []
|
| 43 |
+
all_heights_label = []
|
| 44 |
+
pre_heights_fake = []
|
| 45 |
+
pre_heights_label = []
|
| 46 |
+
mid_heights_fake = []
|
| 47 |
+
mid_heights_label = []
|
| 48 |
+
post_heights_fake = []
|
| 49 |
+
post_heights_label = []
|
| 50 |
+
# 遍历z轴上的每个层
|
| 51 |
+
for z in range(segmentation_label.shape[1]):
|
| 52 |
+
if np.any(segmentation_label[:, z, :]) and np.any(segmentation_fake[:, z, :]):
|
| 53 |
+
segmentation_label_slice = segmentation_label[:, z, :]
|
| 54 |
+
segmentation_fake_slice = segmentation_fake[:, z, :]
|
| 55 |
+
|
| 56 |
+
loc = np.where(segmentation_fake_slice)[1]
|
| 57 |
+
y_min = int(np.min(loc))
|
| 58 |
+
y_max = int(np.max(loc))
|
| 59 |
+
y_range = y_max-y_min
|
| 60 |
+
one_third_y = int(y_min + y_range/3)
|
| 61 |
+
two_third_y = int(y_min + 2*y_range/3)
|
| 62 |
+
center_height_fake = np.count_nonzero(segmentation_fake_slice[:, int(np.mean(loc))])
|
| 63 |
+
all_height_fake = np.count_nonzero(segmentation_fake_slice, axis=0)
|
| 64 |
+
pre_height_fake = np.count_nonzero(segmentation_fake_slice[:,:one_third_y], axis=0)
|
| 65 |
+
mid_height_fake = np.count_nonzero(segmentation_fake_slice[:,one_third_y:two_third_y], axis=0)
|
| 66 |
+
post_height_fake = np.count_nonzero(segmentation_fake_slice[:, two_third_y:], axis=0)
|
| 67 |
+
|
| 68 |
+
loc = np.where(segmentation_label_slice)[1]
|
| 69 |
+
center_height_label = np.count_nonzero(segmentation_label_slice[:, int(np.mean(loc))])
|
| 70 |
+
all_height_label = np.count_nonzero(segmentation_label_slice, axis=0)
|
| 71 |
+
pre_height_label = np.count_nonzero(segmentation_label_slice[:, :one_third_y], axis=0)
|
| 72 |
+
mid_height_label = np.count_nonzero(segmentation_label_slice[:, one_third_y:two_third_y], axis=0)
|
| 73 |
+
post_height_label = np.count_nonzero(segmentation_label_slice[:, two_third_y:], axis=0)
|
| 74 |
+
|
| 75 |
+
|
| 76 |
+
if all_height_label.max()>all_height_fake.max():
|
| 77 |
+
all_scale_ratio = all_height_label.max()/all_height_fake.max()
|
| 78 |
+
else:
|
| 79 |
+
all_scale_ratio = 1
|
| 80 |
+
if pre_height_label.max()>pre_height_fake.max():
|
| 81 |
+
pre_scale_ratio = pre_height_label.max()/pre_height_fake.max()
|
| 82 |
+
else:
|
| 83 |
+
pre_scale_ratio = 1
|
| 84 |
+
if mid_height_label.max()>mid_height_fake.max():
|
| 85 |
+
mid_scale_ratio = mid_height_label.max()/mid_height_fake.max()
|
| 86 |
+
else:
|
| 87 |
+
mid_scale_ratio = 1
|
| 88 |
+
if post_height_label.max()>post_height_fake.max():
|
| 89 |
+
post_scale_ratio = post_height_label.max()/post_height_fake.max()
|
| 90 |
+
else:
|
| 91 |
+
post_scale_ratio = 1
|
| 92 |
+
|
| 93 |
+
all_height_fake = all_height_fake*all_scale_ratio
|
| 94 |
+
center_height_fake = center_height_fake*all_scale_ratio
|
| 95 |
+
pre_height_fake = pre_height_fake*pre_scale_ratio
|
| 96 |
+
mid_height_fake = mid_height_fake*mid_scale_ratio
|
| 97 |
+
post_height_fake = post_height_fake*post_scale_ratio
|
| 98 |
+
|
| 99 |
+
all_heights_fake.extend(all_height_fake[all_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 100 |
+
all_heights_label.extend(all_height_label[all_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 101 |
+
pre_heights_fake.extend(pre_height_fake[pre_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 102 |
+
pre_heights_label.extend(pre_height_label[pre_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 103 |
+
mid_heights_fake.extend(mid_height_fake[mid_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 104 |
+
mid_heights_label.extend(mid_height_label[mid_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 105 |
+
post_heights_fake.extend(post_height_fake[post_height_fake > (center_height_fake*height_threshold)]) # 仅添加非零高度
|
| 106 |
+
post_heights_label.extend(post_height_label[post_height_label > (center_height_label*height_threshold)]) # 仅添加非零高度
|
| 107 |
+
|
| 108 |
+
# 将heights转换为numpy数组以便使用numpy的功能
|
| 109 |
+
all_heights_fake = np.array(all_heights_fake)
|
| 110 |
+
all_heights_label = np.array(all_heights_label)
|
| 111 |
+
pre_heights_fake = np.array(pre_heights_fake)
|
| 112 |
+
pre_heights_label = np.array(pre_heights_label)
|
| 113 |
+
mid_heights_fake = np.array(mid_heights_fake)
|
| 114 |
+
mid_heights_label = np.array(mid_heights_label)
|
| 115 |
+
post_heights_fake = np.array(post_heights_fake)
|
| 116 |
+
post_heights_label = np.array(post_heights_label)
|
| 117 |
+
|
| 118 |
+
return all_heights_fake, all_heights_label,pre_heights_fake, pre_heights_label,mid_heights_fake, mid_heights_label,post_heights_fake, post_heights_label
|
| 119 |
+
|
| 120 |
+
|
| 121 |
+
def calculate_rhlv(segmentation_fake, segmentation_label, center_z, length,vertebra,height_threshold):
|
| 122 |
+
"""
|
| 123 |
+
Calculate the Relative Height Loss Value (RHLV) between fake and label segmentations.
|
| 124 |
+
"""
|
| 125 |
+
seg_fake_filtered = segmentation_fake[:, center_z-length:center_z+length, :]
|
| 126 |
+
seg_label_filtered = segmentation_label[:, center_z-length:center_z+length, :]
|
| 127 |
+
|
| 128 |
+
all_heights_fake, all_heights_label,pre_heights_fake, pre_heights_label,mid_heights_fake, mid_heights_label,post_heights_fake, post_heights_label\
|
| 129 |
+
= calculate_heights(seg_fake_filtered, seg_label_filtered,height_threshold)
|
| 130 |
+
all_height_fake = np.mean(all_heights_fake) if all_heights_fake.size > 0 else 0
|
| 131 |
+
all_height_label = np.mean(all_heights_label) if all_heights_label.size > 0 else 0
|
| 132 |
+
pre_height_fake = np.mean(pre_heights_fake) if pre_heights_fake.size > 0 else 0
|
| 133 |
+
pre_height_label = np.mean(pre_heights_label) if pre_heights_label.size > 0 else 0
|
| 134 |
+
mid_height_fake = np.mean(mid_heights_fake) if mid_heights_fake.size > 0 else 0
|
| 135 |
+
mid_height_label = np.mean(mid_heights_label) if mid_heights_label.size > 0 else 0
|
| 136 |
+
post_height_fake = np.mean(post_heights_fake) if post_heights_fake.size > 0 else 0
|
| 137 |
+
post_height_label = np.mean(post_heights_label) if post_heights_label.size > 0 else 0
|
| 138 |
+
|
| 139 |
+
all_rhlv = (all_height_fake - all_height_label) / (all_height_fake +1e-6)
|
| 140 |
+
pre_rhlv = (pre_height_fake - pre_height_label) / (pre_height_fake +1e-6)
|
| 141 |
+
mid_rhlv = (mid_height_fake - mid_height_label) / (mid_height_fake +1e-6)
|
| 142 |
+
post_rhlv = (post_height_fake - post_height_label) / (post_height_fake +1e-6)
|
| 143 |
+
min_height = min(pre_height_label,mid_height_label,post_height_label)
|
| 144 |
+
max_height = max(pre_height_label,mid_height_label,post_height_label)
|
| 145 |
+
relative_height_label = min_height/(max_height+1e-6)
|
| 146 |
+
|
| 147 |
+
return all_rhlv,pre_rhlv,mid_rhlv,post_rhlv,relative_height_label
|
| 148 |
+
|
| 149 |
+
def process_datasets_to_excel(dataset_info, label_folder, fake_folder, output_file,length_divisor=5, height_threshold=0.64):
|
| 150 |
+
results = []
|
| 151 |
+
for dataset_type, data in dataset_info.items():
|
| 152 |
+
for vertebra, label in data.items():
|
| 153 |
+
label_path = os.path.join(label_folder, vertebra + '.nii.gz')
|
| 154 |
+
fake_path = os.path.join(fake_folder, vertebra + '.nii.gz')
|
| 155 |
+
|
| 156 |
+
if not os.path.exists(label_path) or not os.path.exists(fake_path):
|
| 157 |
+
continue
|
| 158 |
+
|
| 159 |
+
segmentation_label_temp = nib.load(label_path).get_fdata()
|
| 160 |
+
segmentation_label = np.zeros_like(segmentation_label_temp)
|
| 161 |
+
|
| 162 |
+
segmentation_fake_temp = nib.load(fake_path).get_fdata()
|
| 163 |
+
segmentation_fake = np.zeros_like(segmentation_fake_temp)
|
| 164 |
+
|
| 165 |
+
label_index = int(vertebra.split('_')[-1])
|
| 166 |
+
segmentation_label[segmentation_label_temp == label_index] = 1
|
| 167 |
+
segmentation_fake[segmentation_fake_temp == label_index] = 1
|
| 168 |
+
|
| 169 |
+
loc = np.where(segmentation_label)[1]
|
| 170 |
+
if loc.size == 0:
|
| 171 |
+
continue # Skip if no label index found
|
| 172 |
+
|
| 173 |
+
min_z = np.min(loc)
|
| 174 |
+
max_z = np.max(loc)
|
| 175 |
+
center_z = int(np.mean(loc))
|
| 176 |
+
length = (max_z - min_z) // length_divisor # Divisor adjusted based on your setup
|
| 177 |
+
|
| 178 |
+
|
| 179 |
+
all_rhlv, pre_rhlv, mid_rhlv, post_rhlv, relative_height_label = calculate_rhlv(
|
| 180 |
+
segmentation_fake, segmentation_label, center_z, length, vertebra,height_threshold
|
| 181 |
+
)
|
| 182 |
+
print(pre_rhlv,mid_rhlv,post_rhlv)
|
| 183 |
+
results.append({
|
| 184 |
+
"Vertebra": vertebra,
|
| 185 |
+
"Label": label,
|
| 186 |
+
"Dataset": dataset_type,
|
| 187 |
+
"All RHLV": all_rhlv,
|
| 188 |
+
"Pre RHLV": pre_rhlv,
|
| 189 |
+
"Mid RHLV": mid_rhlv,
|
| 190 |
+
"Post RHLV": post_rhlv,
|
| 191 |
+
"Relative Height Label": relative_height_label
|
| 192 |
+
})
|
| 193 |
+
|
| 194 |
+
# Create a DataFrame from results and save to Excel
|
| 195 |
+
df = pd.DataFrame(results)
|
| 196 |
+
df.to_excel(output_file, index=False)
|
| 197 |
+
|
| 198 |
+
def main():
|
| 199 |
+
with open('vertebra_data.json', 'r') as file:
|
| 200 |
+
json_data = json.load(file)
|
| 201 |
+
|
| 202 |
+
label_folder ="datasets/straighten/revised/label"
|
| 203 |
+
output_folder = 'output_3d/coronal'
|
| 204 |
+
result_folder = 'RHLV_quantification'
|
| 205 |
+
if not os.path.exists(result_folder):
|
| 206 |
+
os.makedirs(result_folder)
|
| 207 |
+
for root, dirs, files in os.walk(output_folder):
|
| 208 |
+
for dir in dirs:
|
| 209 |
+
if dir!='fine':
|
| 210 |
+
continue
|
| 211 |
+
|
| 212 |
+
exp_folder = os.path.join(root,dir)
|
| 213 |
+
fake_folder = os.path.join(exp_folder,'label_fake')
|
| 214 |
+
result_file = os.path.join(result_folder,dir+'.xlsx')
|
| 215 |
+
process_datasets_to_excel(json_data, label_folder, fake_folder, result_file, length_divisor=5, height_threshold=0.7)
|
| 216 |
+
|
| 217 |
+
if __name__ == "__main__":
|
| 218 |
+
main()
|
evaluation/SVM_grading.py
ADDED
|
@@ -0,0 +1,96 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
from sklearn.model_selection import StratifiedKFold
|
| 3 |
+
from sklearn.svm import SVC
|
| 4 |
+
from sklearn.metrics import f1_score, precision_score, recall_score, accuracy_score, confusion_matrix
|
| 5 |
+
from sklearn.preprocessing import StandardScaler
|
| 6 |
+
import numpy as np
|
| 7 |
+
import os
|
| 8 |
+
|
| 9 |
+
def evaluate_svm(filepath, features, output_txt='evaluation_results.txt'):
|
| 10 |
+
# 加载数据
|
| 11 |
+
data = pd.read_excel(filepath)
|
| 12 |
+
train_test_data = data[data['Dataset'].isin(['train', 'test'])]
|
| 13 |
+
val_data = data[data['Dataset'] == 'val']
|
| 14 |
+
|
| 15 |
+
# 准备输入和标签
|
| 16 |
+
X_train_test = train_test_data[features]
|
| 17 |
+
y_train_test = train_test_data['Label']
|
| 18 |
+
X_val = val_data[features]
|
| 19 |
+
y_val = val_data['Label']
|
| 20 |
+
|
| 21 |
+
# 数据标准化
|
| 22 |
+
scaler = StandardScaler()
|
| 23 |
+
X_train_test_scaled = scaler.fit_transform(X_train_test)
|
| 24 |
+
X_val_scaled = scaler.transform(X_val)
|
| 25 |
+
|
| 26 |
+
# 初始化 SVM 分类器
|
| 27 |
+
svm_classifier = SVC(kernel='linear', class_weight='balanced')
|
| 28 |
+
|
| 29 |
+
# 设置五折交叉验证
|
| 30 |
+
skf = StratifiedKFold(n_splits=5)
|
| 31 |
+
|
| 32 |
+
# 存储每次验证的结果
|
| 33 |
+
results = []
|
| 34 |
+
f1_list, precision_list, recall_list, accuracy_list = [], [], [], []
|
| 35 |
+
|
| 36 |
+
for train_index, test_index in skf.split(X_train_test_scaled, y_train_test):
|
| 37 |
+
X_train, X_test = X_train_test_scaled[train_index], X_train_test_scaled[test_index]
|
| 38 |
+
y_train, y_test = y_train_test[train_index], y_train_test[test_index]
|
| 39 |
+
|
| 40 |
+
svm_classifier.fit(X_train, y_train)
|
| 41 |
+
y_pred_val = svm_classifier.predict(X_val_scaled)
|
| 42 |
+
cm = confusion_matrix(y_val, y_pred_val)
|
| 43 |
+
f1 = f1_score(y_val, y_pred_val, average='macro')
|
| 44 |
+
precision = precision_score(y_val, y_pred_val, average='macro')
|
| 45 |
+
recall = recall_score(y_val, y_pred_val, average='macro')
|
| 46 |
+
accuracy = accuracy_score(y_val, y_pred_val)
|
| 47 |
+
|
| 48 |
+
results.append((cm, f1, precision, recall, accuracy))
|
| 49 |
+
f1_list.append(f1)
|
| 50 |
+
precision_list.append(precision)
|
| 51 |
+
recall_list.append(recall)
|
| 52 |
+
accuracy_list.append(accuracy)
|
| 53 |
+
|
| 54 |
+
# 写入结果到文件
|
| 55 |
+
with open(output_txt, 'w') as file:
|
| 56 |
+
for i, (cm, f1, precision, recall, accuracy) in enumerate(results):
|
| 57 |
+
file.write(f"Fold {i+1}:\n")
|
| 58 |
+
file.write("Confusion Matrix:\n")
|
| 59 |
+
file.write(f"{cm}\n")
|
| 60 |
+
file.write(f"F1 Score: {f1}, Precision: {precision}, Recall: {recall}, Accuracy: {accuracy}\n")
|
| 61 |
+
file.write("\n")
|
| 62 |
+
|
| 63 |
+
# 计算平均分数和方差
|
| 64 |
+
average_f1 = np.mean(f1_list)
|
| 65 |
+
average_precision = np.mean(precision_list)
|
| 66 |
+
average_recall = np.mean(recall_list)
|
| 67 |
+
average_accuracy = np.mean(accuracy_list)
|
| 68 |
+
variance_f1 = np.var(f1_list)
|
| 69 |
+
variance_precision = np.var(precision_list)
|
| 70 |
+
variance_recall = np.var(recall_list)
|
| 71 |
+
variance_accuracy = np.var(accuracy_list)
|
| 72 |
+
|
| 73 |
+
file.write("Average Scores:\n")
|
| 74 |
+
file.write(f"Average F1 Score: {average_f1} (Variance: {variance_f1})\n")
|
| 75 |
+
file.write(f"Average Precision: {average_precision} (Variance: {variance_precision})\n")
|
| 76 |
+
file.write(f"Average Recall: {average_recall} (Variance: {variance_recall})\n")
|
| 77 |
+
file.write(f"Average Accuracy: {average_accuracy} (Variance: {variance_accuracy})\n")
|
| 78 |
+
|
| 79 |
+
print(f"Results saved to {output_txt}")
|
| 80 |
+
|
| 81 |
+
def main():
|
| 82 |
+
result_folder = 'evaluation/RHLV_quantification'
|
| 83 |
+
grading_folder = 'evaluation/classification_metric'
|
| 84 |
+
if not os.path.exists(grading_folder):
|
| 85 |
+
os.makedirs(grading_folder)
|
| 86 |
+
features = ['Pre RHLV', 'Mid RHLV', 'Post RHLV']
|
| 87 |
+
for xlsx_file in os.listdir(result_folder):
|
| 88 |
+
#if xlsx_file != 'Exp_1_wo_straighten_sagittal.xlsx':
|
| 89 |
+
# continue
|
| 90 |
+
xlsx_path = os.path.join(result_folder, xlsx_file)
|
| 91 |
+
xlsx_name = xlsx_file.split('.')[0]
|
| 92 |
+
saveTxT_path = os.path.join(grading_folder, xlsx_name + '.txt')
|
| 93 |
+
evaluate_svm(xlsx_path, features, saveTxT_path)
|
| 94 |
+
|
| 95 |
+
if __name__ == "__main__":
|
| 96 |
+
main()
|
evaluation/SVM_grading_2.5d.py
ADDED
|
@@ -0,0 +1,100 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
from sklearn.model_selection import StratifiedKFold
|
| 3 |
+
from sklearn.svm import SVC
|
| 4 |
+
from sklearn.metrics import f1_score, precision_score, recall_score, accuracy_score, confusion_matrix
|
| 5 |
+
from sklearn.preprocessing import StandardScaler
|
| 6 |
+
import numpy as np
|
| 7 |
+
import os
|
| 8 |
+
|
| 9 |
+
def evaluate_svm(file1, file2, features, output_txt='evaluation_results.txt'):
|
| 10 |
+
# 加载数据
|
| 11 |
+
data1 = pd.read_excel(file1)
|
| 12 |
+
data2 = pd.read_excel(file2)
|
| 13 |
+
|
| 14 |
+
# 重命名第二个文件的特征,以避免冲突
|
| 15 |
+
rename_dict = {f: f"{f}_2" for f in features}
|
| 16 |
+
data2.rename(columns=rename_dict, inplace=True)
|
| 17 |
+
|
| 18 |
+
# 确保数据有一个共同的列来合并(例如 ID)
|
| 19 |
+
combined_data = pd.merge(data1, data2, on="Vertebra")
|
| 20 |
+
print(combined_data)
|
| 21 |
+
|
| 22 |
+
# 选取参与训练和测试的数据
|
| 23 |
+
train_test_data = combined_data[combined_data['Dataset_x'].isin(['train', 'test'])]
|
| 24 |
+
val_data = combined_data[combined_data['Dataset_x'] == 'val']
|
| 25 |
+
|
| 26 |
+
# 准备输入和标签
|
| 27 |
+
combined_features = features + [f"{f}_2" for f in features]
|
| 28 |
+
X_train_test = train_test_data[combined_features]
|
| 29 |
+
y_train_test = train_test_data['Label_x']
|
| 30 |
+
X_val = val_data[combined_features]
|
| 31 |
+
y_val = val_data['Label_x']
|
| 32 |
+
|
| 33 |
+
# 数据标准化
|
| 34 |
+
scaler = StandardScaler()
|
| 35 |
+
X_train_test_scaled = scaler.fit_transform(X_train_test)
|
| 36 |
+
X_val_scaled = scaler.transform(X_val)
|
| 37 |
+
|
| 38 |
+
# 初始化 SVM 分类器
|
| 39 |
+
svm_classifier = SVC(kernel='linear', class_weight='balanced')
|
| 40 |
+
|
| 41 |
+
# 设置五折交叉验证
|
| 42 |
+
skf = StratifiedKFold(n_splits=5)
|
| 43 |
+
|
| 44 |
+
# 存储每次验证的结果
|
| 45 |
+
results = []
|
| 46 |
+
|
| 47 |
+
for train_index, test_index in skf.split(X_train_test_scaled, y_train_test):
|
| 48 |
+
X_train, X_test = X_train_test_scaled[train_index], X_train_test_scaled[test_index]
|
| 49 |
+
y_train, y_test = y_train_test[train_index], y_train_test[test_index]
|
| 50 |
+
|
| 51 |
+
svm_classifier.fit(X_train, y_train)
|
| 52 |
+
y_pred_val = svm_classifier.predict(X_val_scaled)
|
| 53 |
+
cm = confusion_matrix(y_val, y_pred_val)
|
| 54 |
+
f1 = f1_score(y_val, y_pred_val, average='macro')
|
| 55 |
+
precision = precision_score(y_val, y_pred_val, average='macro')
|
| 56 |
+
recall = recall_score(y_val, y_pred_val, average='macro')
|
| 57 |
+
accuracy = accuracy_score(y_val, y_pred_val)
|
| 58 |
+
|
| 59 |
+
results.append((cm, f1, precision, recall, accuracy))
|
| 60 |
+
|
| 61 |
+
# 写入结果到文件
|
| 62 |
+
with open(output_txt, 'w') as file:
|
| 63 |
+
for i, (cm, f1, precision, recall, accuracy) in enumerate(results):
|
| 64 |
+
file.write(f"Fold {i+1}:\n")
|
| 65 |
+
file.write("Confusion Matrix:\n")
|
| 66 |
+
file.write(f"{cm}\n")
|
| 67 |
+
file.write(f"F1 Score: {f1:.3f}, Precision: {precision:.3f}, Recall: {recall:.3f}, Accuracy: {accuracy:.3f}\n")
|
| 68 |
+
file.write("\n")
|
| 69 |
+
|
| 70 |
+
# 计算平均分数
|
| 71 |
+
average_f1 = np.mean([r[1] for r in results])
|
| 72 |
+
average_precision = np.mean([r[2] for r in results])
|
| 73 |
+
average_recall = np.mean([r[3] for r in results])
|
| 74 |
+
average_accuracy = np.mean([r[4] for r in results])
|
| 75 |
+
|
| 76 |
+
file.write("Average Scores:\n")
|
| 77 |
+
file.write(f"Average F1 Score: {average_f1:.3f}\n")
|
| 78 |
+
file.write(f"Average Precision: {average_precision:.3f}\n")
|
| 79 |
+
file.write(f"Average Recall: {average_recall:.3f}\n")
|
| 80 |
+
file.write(f"Average Accuracy: {average_accuracy:.3f}\n")
|
| 81 |
+
|
| 82 |
+
print(f"Results saved to {output_txt}")
|
| 83 |
+
|
| 84 |
+
def main():
|
| 85 |
+
result_folder = 'RHLV_quantification'
|
| 86 |
+
grading_folder = 'classification_metric'
|
| 87 |
+
if not os.path.exists(grading_folder):
|
| 88 |
+
os.makedirs(grading_folder)
|
| 89 |
+
|
| 90 |
+
file_1 = os.path.join(result_folder,'fine.xlsx')
|
| 91 |
+
#file_1 = 'twostage_output.xlsx'
|
| 92 |
+
file_2 = 'twostage_output.xlsx'
|
| 93 |
+
features = ['Pre RHLV', 'Mid RHLV', 'Post RHLV'] # 特征
|
| 94 |
+
|
| 95 |
+
output_txt_path = os.path.join(grading_folder, 'test.txt')
|
| 96 |
+
evaluate_svm(file_1, file_2, features, output_txt_path)
|
| 97 |
+
|
| 98 |
+
if __name__ == "__main__":
|
| 99 |
+
main()
|
| 100 |
+
|
evaluation/generation_eval_coronal.py
ADDED
|
@@ -0,0 +1,175 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import nibabel as nib
|
| 3 |
+
import numpy as np
|
| 4 |
+
from skimage.metrics import peak_signal_noise_ratio as compare_psnr
|
| 5 |
+
from skimage.metrics import structural_similarity as compare_ssim
|
| 6 |
+
import json
|
| 7 |
+
import pandas as pd
|
| 8 |
+
from sklearn.model_selection import ParameterGrid
|
| 9 |
+
import math
|
| 10 |
+
|
| 11 |
+
def calculate_iou(ori_seg, fake_seg):
|
| 12 |
+
intersection = np.sum(ori_seg * fake_seg)
|
| 13 |
+
union = np.sum(ori_seg + fake_seg > 0)
|
| 14 |
+
if union == 0:
|
| 15 |
+
return 0
|
| 16 |
+
else:
|
| 17 |
+
return intersection / union
|
| 18 |
+
|
| 19 |
+
def calculate_dice(ori_seg, fake_seg):
|
| 20 |
+
# 计算两个分割之间的交集
|
| 21 |
+
intersection = np.sum(ori_seg * fake_seg)
|
| 22 |
+
# 计算两个分割之间的并集
|
| 23 |
+
union = np.sum(ori_seg) + np.sum(fake_seg)
|
| 24 |
+
# 如果并集为零,返回0,否则返回Dice系数
|
| 25 |
+
if union == 0:
|
| 26 |
+
return 0
|
| 27 |
+
else:
|
| 28 |
+
return 2.0 * intersection / union
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
def relative_volume_difference(ori_seg, fake_seg):
|
| 32 |
+
volume_ori = np.sum(ori_seg)
|
| 33 |
+
volume_fake = np.sum(fake_seg)
|
| 34 |
+
if volume_ori == 0:
|
| 35 |
+
return 0
|
| 36 |
+
else:
|
| 37 |
+
return np.abs(volume_ori - volume_fake) / volume_ori
|
| 38 |
+
|
| 39 |
+
def process_images(ori_ct_path, fake_ct_path, ori_seg_path, fake_seg_path):
|
| 40 |
+
ori_ct = nib.load(ori_ct_path).get_fdata()
|
| 41 |
+
fake_ct = nib.load(fake_ct_path).get_fdata()
|
| 42 |
+
ori_seg_temp = nib.load(ori_seg_path).get_fdata()
|
| 43 |
+
ori_seg = np.zeros_like(ori_seg_temp)
|
| 44 |
+
fake_seg_temp = nib.load(fake_seg_path).get_fdata()
|
| 45 |
+
fake_seg = np.zeros_like(fake_seg_temp)
|
| 46 |
+
|
| 47 |
+
label = int(ori_seg_path[:-7].split('_')[-1])
|
| 48 |
+
ori_seg[ori_seg_temp==label] = 1
|
| 49 |
+
fake_seg[fake_seg_temp==label] = 1
|
| 50 |
+
|
| 51 |
+
patch_psnr_list = []
|
| 52 |
+
patch_ssim_list = []
|
| 53 |
+
global_psnr_list = []
|
| 54 |
+
global_ssim_list = []
|
| 55 |
+
|
| 56 |
+
iou_value = calculate_iou(ori_seg, fake_seg)
|
| 57 |
+
dice_value = calculate_dice(ori_seg, fake_seg)
|
| 58 |
+
rv_diff = relative_volume_difference(ori_seg, fake_seg)
|
| 59 |
+
|
| 60 |
+
loc = np.where(ori_seg)
|
| 61 |
+
z0 = min(loc[1])
|
| 62 |
+
z1 = max(loc[1])
|
| 63 |
+
range_length = z1 - z0 + 1
|
| 64 |
+
new_range_length = int(range_length * 4 / 5)
|
| 65 |
+
new_z0 = z0 + (range_length - new_range_length) // 2
|
| 66 |
+
new_z1 = new_z0 + new_range_length - 1
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
|
| 70 |
+
for z in range(new_z0, new_z1 + 1):
|
| 71 |
+
if np.sum(ori_seg[:,z,:]) > 400:
|
| 72 |
+
coords = np.argwhere(ori_seg[:,z,:])
|
| 73 |
+
x1, x2 = min(coords[:, 0]), max(coords[:, 0])
|
| 74 |
+
|
| 75 |
+
crop_ori_ct = ori_ct[x1:x2+1, z, :]
|
| 76 |
+
crop_fake_ct = fake_ct[x1:x2+1, z, :]
|
| 77 |
+
|
| 78 |
+
psnr_value = compare_psnr(crop_ori_ct, crop_fake_ct, data_range=crop_ori_ct.max() - crop_ori_ct.min())
|
| 79 |
+
ssim_value = compare_ssim(crop_ori_ct, crop_fake_ct, data_range=crop_ori_ct.max() - crop_ori_ct.min())
|
| 80 |
+
|
| 81 |
+
if not np.isnan(psnr_value):
|
| 82 |
+
patch_psnr_list.append(psnr_value)
|
| 83 |
+
if not np.isnan(ssim_value):
|
| 84 |
+
patch_ssim_list.append(ssim_value)
|
| 85 |
+
|
| 86 |
+
for z in range(new_z0, new_z1 + 1):
|
| 87 |
+
if np.sum(ori_seg[:,z,:]) > 400:
|
| 88 |
+
psnr_value = compare_psnr(ori_ct[:,z,:], fake_ct[:,z,:], data_range=ori_ct[:,z,:].max() - ori_ct[:,z,:].min())
|
| 89 |
+
ssim_value = compare_ssim(ori_ct[:,z,:], fake_ct[:,z,:], data_range=ori_ct[:,z,:].max() - ori_ct[:,z,:].min())
|
| 90 |
+
|
| 91 |
+
if not np.isnan(psnr_value):
|
| 92 |
+
global_psnr_list.append(psnr_value)
|
| 93 |
+
if not np.isnan(ssim_value):
|
| 94 |
+
global_ssim_list.append(ssim_value)
|
| 95 |
+
|
| 96 |
+
avg_patch_psnr = np.mean(patch_psnr_list) if patch_psnr_list else 0 # 检查列表是否为空
|
| 97 |
+
avg_patch_ssim = np.mean(patch_ssim_list) if patch_ssim_list else 0 # 检查列表是否为空
|
| 98 |
+
avg_global_psnr = np.mean(global_psnr_list) if global_psnr_list else 0 # 检查列表是否为空
|
| 99 |
+
avg_global_ssim = np.mean(global_ssim_list) if global_ssim_list else 0 # 检查列表是否为空
|
| 100 |
+
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
return avg_global_psnr, avg_global_ssim, avg_patch_psnr, avg_patch_ssim, iou_value, rv_diff, dice_value
|
| 104 |
+
|
| 105 |
+
def average_metrics(lists):
|
| 106 |
+
return np.mean(lists)
|
| 107 |
+
|
| 108 |
+
def main():
|
| 109 |
+
ori_ct_folder = '/home/ubuntu/Project/HealthiVert-GAN/datasets/straighten/CT'
|
| 110 |
+
ori_seg_folder = '/home/ubuntu/Project/HealthiVert-GAN/datasets/straighten/label'
|
| 111 |
+
json_path = 'vertebra_data.json'
|
| 112 |
+
save_folder = "evaluation/generation_metric"
|
| 113 |
+
output_folder = '/home/ubuntu/Project/HealthiVert-GAN_eval/output'
|
| 114 |
+
with open(json_path, 'r') as file:
|
| 115 |
+
vertebra_set = json.load(file)
|
| 116 |
+
val_normal_vert = []
|
| 117 |
+
for patient_vert_id in vertebra_set['val'].keys():
|
| 118 |
+
if int(vertebra_set['val'][patient_vert_id]) == 0:
|
| 119 |
+
val_normal_vert.append(patient_vert_id)
|
| 120 |
+
|
| 121 |
+
if not os.path.exists(save_folder):
|
| 122 |
+
os.makedirs(save_folder)
|
| 123 |
+
|
| 124 |
+
first_level_directories = []
|
| 125 |
+
for root, dirs, files in os.walk(output_folder):
|
| 126 |
+
first_level_directories.extend([os.path.join(root, d) for d in dirs])
|
| 127 |
+
break # 退出循环以避免进入更深层次的目录
|
| 128 |
+
print(first_level_directories)
|
| 129 |
+
|
| 130 |
+
for root, dirs, files in os.walk(output_folder):
|
| 131 |
+
for dir in dirs:
|
| 132 |
+
#if dir != 'Exp_3_mask3ver':
|
| 133 |
+
# continue
|
| 134 |
+
if 'coronal' not in dir:
|
| 135 |
+
continue
|
| 136 |
+
exp_folder = os.path.join(root,dir)
|
| 137 |
+
fake_seg_folder = os.path.join(exp_folder,'label_fake')
|
| 138 |
+
fake_ct_folder = os.path.join(exp_folder,'CT_fake')
|
| 139 |
+
|
| 140 |
+
metrics_lists = {'global_psnr': [], 'global_ssim': [], 'patch_psnr': [], 'patch_ssim': [], 'iou': [], 'rv_diff': [], 'dice':[]}
|
| 141 |
+
count=0
|
| 142 |
+
for filename in os.listdir(ori_ct_folder):
|
| 143 |
+
|
| 144 |
+
if filename.endswith(".nii.gz") and filename[:-7] in val_normal_vert:
|
| 145 |
+
ori_ct_path = os.path.join(ori_ct_folder, filename)
|
| 146 |
+
fake_ct_path = os.path.join(fake_ct_folder, filename)
|
| 147 |
+
ori_seg_path = os.path.join(ori_seg_folder, filename)
|
| 148 |
+
fake_seg_path = os.path.join(fake_seg_folder, filename)
|
| 149 |
+
|
| 150 |
+
global_psnr, global_ssim, patch_psnr, patch_ssim, iou, rv_diff, dice = process_images(
|
| 151 |
+
ori_ct_path, fake_ct_path, ori_seg_path, fake_seg_path)
|
| 152 |
+
if math.isnan(patch_psnr) or math.isnan(patch_ssim):
|
| 153 |
+
print("PSNR or SSIM returned NaN, skipping this set of images.")
|
| 154 |
+
continue
|
| 155 |
+
if patch_psnr==0 or patch_ssim==0:
|
| 156 |
+
print("PSNR or SSIM returned 0, skipping this set of images.")
|
| 157 |
+
continue
|
| 158 |
+
metrics_lists['global_psnr'].append(global_psnr)
|
| 159 |
+
metrics_lists['global_ssim'].append(global_ssim)
|
| 160 |
+
metrics_lists['patch_psnr'].append(patch_psnr)
|
| 161 |
+
metrics_lists['patch_ssim'].append(patch_ssim)
|
| 162 |
+
metrics_lists['iou'].append(iou)
|
| 163 |
+
metrics_lists['rv_diff'].append(rv_diff)
|
| 164 |
+
metrics_lists['dice'].append(dice)
|
| 165 |
+
count+=1
|
| 166 |
+
|
| 167 |
+
# 计算总平均
|
| 168 |
+
avg_metrics = {key: average_metrics(value) for key, value in metrics_lists.items()}
|
| 169 |
+
|
| 170 |
+
with open(os.path.join(save_folder,dir+".txt"), "w") as file:
|
| 171 |
+
for metric, value in avg_metrics.items():
|
| 172 |
+
file.write(f"Average {metric.upper()}: {value}\n")
|
| 173 |
+
|
| 174 |
+
if __name__ == "__main__":
|
| 175 |
+
main()
|
evaluation/generation_eval_sagittal.py
ADDED
|
@@ -0,0 +1,165 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import nibabel as nib
|
| 3 |
+
import numpy as np
|
| 4 |
+
from skimage.metrics import peak_signal_noise_ratio as compare_psnr
|
| 5 |
+
from skimage.metrics import structural_similarity as compare_ssim
|
| 6 |
+
import json
|
| 7 |
+
import pandas as pd
|
| 8 |
+
from sklearn.model_selection import ParameterGrid
|
| 9 |
+
import math
|
| 10 |
+
|
| 11 |
+
def calculate_iou(ori_seg, fake_seg):
|
| 12 |
+
intersection = np.sum(ori_seg * fake_seg)
|
| 13 |
+
union = np.sum(ori_seg + fake_seg > 0)
|
| 14 |
+
if union == 0:
|
| 15 |
+
return 0
|
| 16 |
+
else:
|
| 17 |
+
return intersection / union
|
| 18 |
+
|
| 19 |
+
def calculate_dice(ori_seg, fake_seg):
|
| 20 |
+
# 计算两个分割之间的交集
|
| 21 |
+
intersection = np.sum(ori_seg * fake_seg)
|
| 22 |
+
# 计算两个分割之间的并集
|
| 23 |
+
union = np.sum(ori_seg) + np.sum(fake_seg)
|
| 24 |
+
# 如果并集为零,返回0,否则返回Dice系数
|
| 25 |
+
if union == 0:
|
| 26 |
+
return 0
|
| 27 |
+
else:
|
| 28 |
+
return 2.0 * intersection / union
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
def relative_volume_difference(ori_seg, fake_seg):
|
| 32 |
+
volume_ori = np.sum(ori_seg)
|
| 33 |
+
volume_fake = np.sum(fake_seg)
|
| 34 |
+
if volume_ori == 0:
|
| 35 |
+
return 0
|
| 36 |
+
else:
|
| 37 |
+
return np.abs(volume_ori - volume_fake) / volume_ori
|
| 38 |
+
|
| 39 |
+
def process_images(ori_ct_path, fake_ct_path, ori_seg_path, fake_seg_path):
|
| 40 |
+
ori_ct = nib.load(ori_ct_path).get_fdata()
|
| 41 |
+
fake_ct = nib.load(fake_ct_path).get_fdata()
|
| 42 |
+
ori_seg_temp = nib.load(ori_seg_path).get_fdata()
|
| 43 |
+
ori_seg = np.zeros_like(ori_seg_temp)
|
| 44 |
+
fake_seg_temp = nib.load(fake_seg_path).get_fdata()
|
| 45 |
+
fake_seg = np.zeros_like(fake_seg_temp)
|
| 46 |
+
|
| 47 |
+
label = int(ori_seg_path[:-7].split('_')[-1])
|
| 48 |
+
ori_seg[ori_seg_temp==label] = 1
|
| 49 |
+
fake_seg[fake_seg_temp==label] = 1
|
| 50 |
+
|
| 51 |
+
patch_psnr_list = []
|
| 52 |
+
patch_ssim_list = []
|
| 53 |
+
global_psnr_list = []
|
| 54 |
+
global_ssim_list = []
|
| 55 |
+
|
| 56 |
+
iou_value = calculate_iou(ori_seg, fake_seg)
|
| 57 |
+
dice_value = calculate_dice(ori_seg, fake_seg)
|
| 58 |
+
rv_diff = relative_volume_difference(ori_seg, fake_seg)
|
| 59 |
+
|
| 60 |
+
loc = np.where(ori_seg)
|
| 61 |
+
z0 = min(loc[2])
|
| 62 |
+
z1 = max(loc[2])
|
| 63 |
+
range_length = z1 - z0 + 1
|
| 64 |
+
new_range_length = int(range_length * 4 / 5)
|
| 65 |
+
new_z0 = z0 + (range_length - new_range_length) // 2
|
| 66 |
+
new_z1 = new_z0 + new_range_length - 1
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
|
| 70 |
+
for z in range(new_z0, new_z1 + 1):
|
| 71 |
+
if np.sum(ori_seg[:,:,z]) > 400:
|
| 72 |
+
coords = np.argwhere(ori_seg[:,:,z])
|
| 73 |
+
x1, x2 = min(coords[:, 0]), max(coords[:, 0])
|
| 74 |
+
|
| 75 |
+
crop_ori_ct = ori_ct[x1:x2+1, :, z]
|
| 76 |
+
crop_fake_ct = fake_ct[x1:x2+1, :, z]
|
| 77 |
+
|
| 78 |
+
psnr_value = compare_psnr(crop_ori_ct, crop_fake_ct, data_range=crop_ori_ct.max() - crop_ori_ct.min())
|
| 79 |
+
ssim_value = compare_ssim(crop_ori_ct, crop_fake_ct, data_range=crop_ori_ct.max() - crop_ori_ct.min())
|
| 80 |
+
|
| 81 |
+
if not np.isnan(psnr_value):
|
| 82 |
+
patch_psnr_list.append(psnr_value)
|
| 83 |
+
if not np.isnan(ssim_value):
|
| 84 |
+
patch_ssim_list.append(ssim_value)
|
| 85 |
+
|
| 86 |
+
for z in range(new_z0, new_z1 + 1):
|
| 87 |
+
if np.sum(ori_seg[:,:,z]) > 400:
|
| 88 |
+
psnr_value = compare_psnr(ori_ct[:,:,z], fake_ct[:,:,z], data_range=ori_ct[:,:,z].max() - ori_ct[:,:,z].min())
|
| 89 |
+
ssim_value = compare_ssim(ori_ct[:,:,z], fake_ct[:,:,z], data_range=ori_ct[:,:,z].max() - ori_ct[:,:,z].min())
|
| 90 |
+
|
| 91 |
+
if not np.isnan(psnr_value):
|
| 92 |
+
global_psnr_list.append(psnr_value)
|
| 93 |
+
if not np.isnan(ssim_value):
|
| 94 |
+
global_ssim_list.append(ssim_value)
|
| 95 |
+
|
| 96 |
+
avg_patch_psnr = np.mean(patch_psnr_list) if patch_psnr_list else 0 # 检查列表是否为空
|
| 97 |
+
avg_patch_ssim = np.mean(patch_ssim_list) if patch_ssim_list else 0 # 检查列表是否为空
|
| 98 |
+
avg_global_psnr = np.mean(global_psnr_list) if global_psnr_list else 0 # 检查列表是否为空
|
| 99 |
+
avg_global_ssim = np.mean(global_ssim_list) if global_ssim_list else 0 # 检查列表是否为空
|
| 100 |
+
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
return avg_global_psnr, avg_global_ssim, avg_patch_psnr, avg_patch_ssim, iou_value, rv_diff, dice_value
|
| 104 |
+
|
| 105 |
+
def average_metrics(lists):
|
| 106 |
+
return np.mean(lists)
|
| 107 |
+
|
| 108 |
+
def main():
|
| 109 |
+
ori_ct_folder = '/dssg/home/acct-milesun/zhangqi/Dataset/HealthiVert_straighten/CT'
|
| 110 |
+
ori_seg_folder = '/dssg/home/acct-milesun/zhangqi/Dataset/HealthiVert_straighten/label'
|
| 111 |
+
json_path = 'vertebra_data.json'
|
| 112 |
+
save_folder = "evaluation/generation_metric"
|
| 113 |
+
output_folder = '/dssg/home/acct-milesun/zhangqi/Project/HealthiVert-GAN_eval/output'
|
| 114 |
+
with open(json_path, 'r') as file:
|
| 115 |
+
vertebra_set = json.load(file)
|
| 116 |
+
val_normal_vert = []
|
| 117 |
+
for patient_vert_id in vertebra_set['val'].keys():
|
| 118 |
+
if int(vertebra_set['val'][patient_vert_id]) == 0:
|
| 119 |
+
val_normal_vert.append(patient_vert_id)
|
| 120 |
+
|
| 121 |
+
if not os.path.exists(save_folder):
|
| 122 |
+
os.makedirs(save_folder)
|
| 123 |
+
|
| 124 |
+
for root, dirs, files in os.walk(output_folder):
|
| 125 |
+
for dir in dirs:
|
| 126 |
+
exp_folder = os.path.join(root,dir)
|
| 127 |
+
fake_seg_folder = os.path.join(exp_folder,'label_fake')
|
| 128 |
+
fake_ct_folder = os.path.join(exp_folder,'CT_fake')
|
| 129 |
+
|
| 130 |
+
metrics_lists = {'global_psnr': [], 'global_ssim': [], 'patch_psnr': [], 'patch_ssim': [], 'iou': [], 'rv_diff': [], 'dice':[]}
|
| 131 |
+
count=0
|
| 132 |
+
for filename in os.listdir(ori_ct_folder):
|
| 133 |
+
|
| 134 |
+
if filename.endswith(".nii.gz") and filename[:-7] in val_normal_vert:
|
| 135 |
+
ori_ct_path = os.path.join(ori_ct_folder, filename)
|
| 136 |
+
fake_ct_path = os.path.join(fake_ct_folder, filename)
|
| 137 |
+
ori_seg_path = os.path.join(ori_seg_folder, filename)
|
| 138 |
+
fake_seg_path = os.path.join(fake_seg_folder, filename)
|
| 139 |
+
|
| 140 |
+
global_psnr, global_ssim, patch_psnr, patch_ssim, iou, rv_diff, dice = process_images(
|
| 141 |
+
ori_ct_path, fake_ct_path, ori_seg_path, fake_seg_path)
|
| 142 |
+
if math.isnan(patch_psnr) or math.isnan(patch_ssim):
|
| 143 |
+
print("PSNR or SSIM returned NaN, skipping this set of images.")
|
| 144 |
+
continue
|
| 145 |
+
if patch_psnr==0 or patch_ssim==0:
|
| 146 |
+
print("PSNR or SSIM returned 0, skipping this set of images.")
|
| 147 |
+
continue
|
| 148 |
+
metrics_lists['global_psnr'].append(global_psnr)
|
| 149 |
+
metrics_lists['global_ssim'].append(global_ssim)
|
| 150 |
+
metrics_lists['patch_psnr'].append(patch_psnr)
|
| 151 |
+
metrics_lists['patch_ssim'].append(patch_ssim)
|
| 152 |
+
metrics_lists['iou'].append(iou)
|
| 153 |
+
metrics_lists['rv_diff'].append(rv_diff)
|
| 154 |
+
metrics_lists['dice'].append(dice)
|
| 155 |
+
count+=1
|
| 156 |
+
|
| 157 |
+
# 计算总平均
|
| 158 |
+
avg_metrics = {key: average_metrics(value) for key, value in metrics_lists.items()}
|
| 159 |
+
|
| 160 |
+
with open(os.path.join(save_folder,dir+".txt"), "w") as file:
|
| 161 |
+
for metric, value in avg_metrics.items():
|
| 162 |
+
file.write(f"Average {metric.upper()}: {value}\n")
|
| 163 |
+
|
| 164 |
+
if __name__ == "__main__":
|
| 165 |
+
main()
|
images/SHRM_and_HGAM.png
ADDED

|
Git LFS Details
|
images/attention.png
ADDED

|
Git LFS Details
|
images/comparison_with_others.png
ADDED

|
Git LFS Details
|
images/distribution.png
ADDED

|
Git LFS Details
|
images/mask.png
ADDED

|
Git LFS Details
|
images/network.png
ADDED

|
Git LFS Details
|
images/our_method.png
ADDED

|
Git LFS Details
|