Spaces:
Running
Running
thomas0809
commited on
Commit
·
bec3c91
1
Parent(s):
80e0f28
add indigo render
Browse files
app.py
CHANGED
|
@@ -2,8 +2,12 @@ import gradio as gr
|
|
| 2 |
|
| 3 |
import os
|
| 4 |
import glob
|
|
|
|
|
|
|
| 5 |
import torch
|
| 6 |
from molscribe import MolScribe
|
|
|
|
|
|
|
| 7 |
|
| 8 |
from huggingface_hub import hf_hub_download
|
| 9 |
|
|
@@ -14,21 +18,38 @@ ckpt_path = hf_hub_download(REPO_ID, FILENAME)
|
|
| 14 |
device = torch.device('cpu')
|
| 15 |
model = MolScribe(ckpt_path, device)
|
| 16 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 17 |
def predict(image):
|
| 18 |
smiles, molblock = model.predict_image(image)
|
| 19 |
-
|
|
|
|
| 20 |
|
| 21 |
iface = gr.Interface(
|
| 22 |
predict,
|
| 23 |
-
inputs=gr.Image(label="Upload molecular image"),
|
| 24 |
outputs=[
|
|
|
|
| 25 |
gr.Textbox(label="SMILES"),
|
| 26 |
gr.Textbox(label="Molfile"),
|
| 27 |
],
|
| 28 |
allow_flagging="auto",
|
| 29 |
title="MolScribe",
|
| 30 |
-
description="Convert a molecular image into SMILES and Molfile.
|
|
|
|
|
|
|
|
|
|
| 31 |
examples=sorted(glob.glob('examples/*.png')),
|
| 32 |
examples_per_page=20,
|
| 33 |
)
|
| 34 |
-
iface.launch()
|
|
|
|
| 2 |
|
| 3 |
import os
|
| 4 |
import glob
|
| 5 |
+
import cv2
|
| 6 |
+
import numpy as np
|
| 7 |
import torch
|
| 8 |
from molscribe import MolScribe
|
| 9 |
+
from indigo import Indigo
|
| 10 |
+
from indigo.renderer import IndigoRenderer
|
| 11 |
|
| 12 |
from huggingface_hub import hf_hub_download
|
| 13 |
|
|
|
|
| 18 |
device = torch.device('cpu')
|
| 19 |
model = MolScribe(ckpt_path, device)
|
| 20 |
|
| 21 |
+
def generate_image(molblock):
|
| 22 |
+
indigo = Indigo()
|
| 23 |
+
render = IndigoRenderer(indigo)
|
| 24 |
+
indigo.setOption('render-output-format', 'png')
|
| 25 |
+
indigo.setOption('render-background-color', '1,1,1')
|
| 26 |
+
indigo.setOption('render-stereo-style', 'none')
|
| 27 |
+
indigo.setOption('render-label-mode', 'hetero')
|
| 28 |
+
mol = indigo.loadMolecule(molblock)
|
| 29 |
+
buf = render.renderToBuffer(mol)
|
| 30 |
+
img = cv2.imdecode(np.asarray(bytearray(buf), dtype=np.uint8), 1)
|
| 31 |
+
return img
|
| 32 |
+
|
| 33 |
def predict(image):
|
| 34 |
smiles, molblock = model.predict_image(image)
|
| 35 |
+
image = generate_image(molblock)
|
| 36 |
+
return image, smiles, molblock
|
| 37 |
|
| 38 |
iface = gr.Interface(
|
| 39 |
predict,
|
| 40 |
+
inputs=gr.Image(label="Upload molecular image", show_label=False).style(height=256),
|
| 41 |
outputs=[
|
| 42 |
+
gr.Image(label="Prediction").style(height=256),
|
| 43 |
gr.Textbox(label="SMILES"),
|
| 44 |
gr.Textbox(label="Molfile"),
|
| 45 |
],
|
| 46 |
allow_flagging="auto",
|
| 47 |
title="MolScribe",
|
| 48 |
+
description="Convert a molecular image into SMILES and Molfile. (To view the prediction better, copy-paste the Molfile to ChemDraw.) <br> " \
|
| 49 |
+
"Paper: [_MolScribe: Robust Molecular Structure Recognition with Image-To-Graph Generation_](https://arxiv.org/abs/2205.14311) <br>" \
|
| 50 |
+
"Code: https://github.com/thomas0809/MolScribe <br>" \
|
| 51 |
+
"Authors: [Yujie Qian](mailto:[email protected]), Jiang Guo, Zhengkai Tu, Zhening Li, Connor W. Coley, Regina Barzilay. _MIT CSAIL_.",
|
| 52 |
examples=sorted(glob.glob('examples/*.png')),
|
| 53 |
examples_per_page=20,
|
| 54 |
)
|
| 55 |
+
iface.launch()
|
examples/US20040186132A1_p0003_x0634_y1617_c00000.png
ADDED
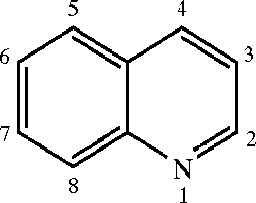
|
examples/US20040229890A1_p0002_x1627_y1019_c00002.png
ADDED
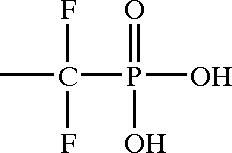
|
examples/US20040229890A1_p0014_x0525_y2643_c00061.png
ADDED

|
examples/US20040266789A1_p0035_x1301_y2162_c00060.png
ADDED
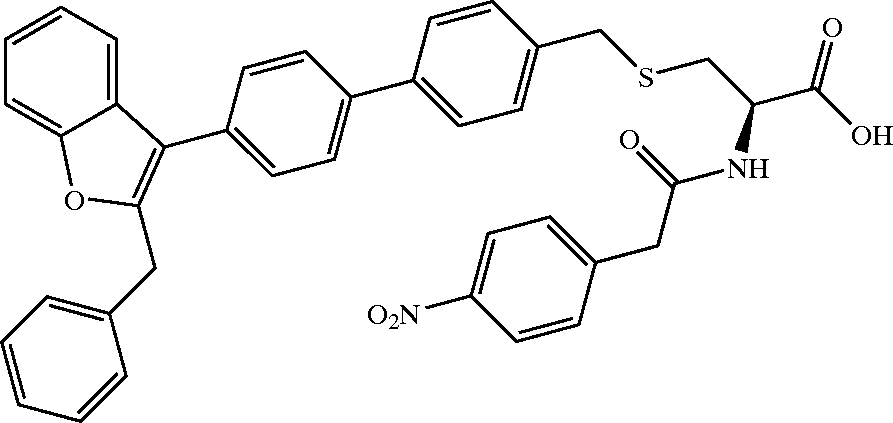
|
requirements.txt
CHANGED
|
@@ -1 +1,2 @@
|
|
| 1 |
git+https://github.com/thomas0809/MolScribe.git@main#egg=MolScribe
|
|
|
|
|
|
| 1 |
git+https://github.com/thomas0809/MolScribe.git@main#egg=MolScribe
|
| 2 |
+
epam.indigo==1.8.0
|