Commit
·
d05f89f
1
Parent(s):
99d658f
Synced repo using 'sync_with_huggingface' Github Action
Browse files- .pre-commit-config.yaml +13 -0
- LICENSE +674 -0
- MANIFEST.in +3 -0
- b3clf/BBB_general_workflow_v4.png +0 -0
- b3clf/__init__.py +31 -0
- b3clf/__main__.py +109 -0
- b3clf/b3clf.py +154 -0
- b3clf/b3clf_structure.png +0 -0
- b3clf/data/B3clf_thresholds.xlsx +0 -0
- b3clf/descriptor_padel.py +90 -0
- b3clf/feature_list.txt +475 -0
- b3clf/geometry_opt.py +208 -0
- b3clf/pre_trained/b3clf_dtree_borderline_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_dtree_classic_ADASYN.joblib +3 -0
- b3clf/pre_trained/b3clf_dtree_classic_RandUndersampling.joblib +3 -0
- b3clf/pre_trained/b3clf_dtree_classic_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_dtree_common.joblib +3 -0
- b3clf/pre_trained/b3clf_dtree_kmeans_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_borderline_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_classic_ADASYN.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_classic_RandUndersampling.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_classic_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_common.joblib +3 -0
- b3clf/pre_trained/b3clf_knn_kmeans_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_borderline_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_classic_ADASYN.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_classic_RandUndersampling.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_classic_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_common.joblib +3 -0
- b3clf/pre_trained/b3clf_logreg_kmeans_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_scaler.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_borderline_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_classic_ADASYN.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_classic_RandUndersampling.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_classic_SMOTE.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_common.joblib +3 -0
- b3clf/pre_trained/b3clf_xgb_kmeans_SMOTE.joblib +3 -0
- b3clf/test/test_SMILES.csv +7 -0
- b3clf/test/test_input_sdf.sdf +387 -0
- b3clf/test/test_padel_descriptors.xlsx +0 -0
- b3clf/utils.py +161 -0
- b3clf/version.py +29 -0
- requirements.txt +9 -0
- requirements_conda.txt +9 -0
- setup.py +83 -0
.pre-commit-config.yaml
ADDED
|
@@ -0,0 +1,13 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
minimum_pre_commit_version: 3.4.0
|
| 2 |
+
repos:
|
| 3 |
+
- repo: https://github.com/pre-commit/pre-commit-hooks
|
| 4 |
+
rev: v4.4.0
|
| 5 |
+
hooks:
|
| 6 |
+
- id: check-merge-conflict # Check for files that contain merge conflict strings.
|
| 7 |
+
- id: trailing-whitespace # Trims trailing whitespace.
|
| 8 |
+
args: [--markdown-linebreak-ext=md]
|
| 9 |
+
- id: mixed-line-ending # Replaces or checks mixed line ending.
|
| 10 |
+
args: [--fix=lf]
|
| 11 |
+
- id: end-of-file-fixer # Makes sure files end in a newline and only a newline.
|
| 12 |
+
- id: check-merge-conflict # Check for files that contain merge conflict strings.
|
| 13 |
+
- id: check-ast # Simply check whether files parse as valid python.
|
LICENSE
ADDED
|
@@ -0,0 +1,674 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
GNU GENERAL PUBLIC LICENSE
|
| 2 |
+
Version 3, 29 June 2007
|
| 3 |
+
|
| 4 |
+
Copyright (C) 2007 Free Software Foundation, Inc. <http://fsf.org/>
|
| 5 |
+
Everyone is permitted to copy and distribute verbatim copies
|
| 6 |
+
of this license document, but changing it is not allowed.
|
| 7 |
+
|
| 8 |
+
Preamble
|
| 9 |
+
|
| 10 |
+
The GNU General Public License is a free, copyleft license for
|
| 11 |
+
software and other kinds of works.
|
| 12 |
+
|
| 13 |
+
The licenses for most software and other practical works are designed
|
| 14 |
+
to take away your freedom to share and change the works. By contrast,
|
| 15 |
+
the GNU General Public License is intended to guarantee your freedom to
|
| 16 |
+
share and change all versions of a program--to make sure it remains free
|
| 17 |
+
software for all its users. We, the Free Software Foundation, use the
|
| 18 |
+
GNU General Public License for most of our software; it applies also to
|
| 19 |
+
any other work released this way by its authors. You can apply it to
|
| 20 |
+
your programs, too.
|
| 21 |
+
|
| 22 |
+
When we speak of free software, we are referring to freedom, not
|
| 23 |
+
price. Our General Public Licenses are designed to make sure that you
|
| 24 |
+
have the freedom to distribute copies of free software (and charge for
|
| 25 |
+
them if you wish), that you receive source code or can get it if you
|
| 26 |
+
want it, that you can change the software or use pieces of it in new
|
| 27 |
+
free programs, and that you know you can do these things.
|
| 28 |
+
|
| 29 |
+
To protect your rights, we need to prevent others from denying you
|
| 30 |
+
these rights or asking you to surrender the rights. Therefore, you have
|
| 31 |
+
certain responsibilities if you distribute copies of the software, or if
|
| 32 |
+
you modify it: responsibilities to respect the freedom of others.
|
| 33 |
+
|
| 34 |
+
For example, if you distribute copies of such a program, whether
|
| 35 |
+
gratis or for a fee, you must pass on to the recipients the same
|
| 36 |
+
freedoms that you received. You must make sure that they, too, receive
|
| 37 |
+
or can get the source code. And you must show them these terms so they
|
| 38 |
+
know their rights.
|
| 39 |
+
|
| 40 |
+
Developers that use the GNU GPL protect your rights with two steps:
|
| 41 |
+
(1) assert copyright on the software, and (2) offer you this License
|
| 42 |
+
giving you legal permission to copy, distribute and/or modify it.
|
| 43 |
+
|
| 44 |
+
For the developers' and authors' protection, the GPL clearly explains
|
| 45 |
+
that there is no warranty for this free software. For both users' and
|
| 46 |
+
authors' sake, the GPL requires that modified versions be marked as
|
| 47 |
+
changed, so that their problems will not be attributed erroneously to
|
| 48 |
+
authors of previous versions.
|
| 49 |
+
|
| 50 |
+
Some devices are designed to deny users access to install or run
|
| 51 |
+
modified versions of the software inside them, although the manufacturer
|
| 52 |
+
can do so. This is fundamentally incompatible with the aim of
|
| 53 |
+
protecting users' freedom to change the software. The systematic
|
| 54 |
+
pattern of such abuse occurs in the area of products for individuals to
|
| 55 |
+
use, which is precisely where it is most unacceptable. Therefore, we
|
| 56 |
+
have designed this version of the GPL to prohibit the practice for those
|
| 57 |
+
products. If such problems arise substantially in other domains, we
|
| 58 |
+
stand ready to extend this provision to those domains in future versions
|
| 59 |
+
of the GPL, as needed to protect the freedom of users.
|
| 60 |
+
|
| 61 |
+
Finally, every program is threatened constantly by software patents.
|
| 62 |
+
States should not allow patents to restrict development and use of
|
| 63 |
+
software on general-purpose computers, but in those that do, we wish to
|
| 64 |
+
avoid the special danger that patents applied to a free program could
|
| 65 |
+
make it effectively proprietary. To prevent this, the GPL assures that
|
| 66 |
+
patents cannot be used to render the program non-free.
|
| 67 |
+
|
| 68 |
+
The precise terms and conditions for copying, distribution and
|
| 69 |
+
modification follow.
|
| 70 |
+
|
| 71 |
+
TERMS AND CONDITIONS
|
| 72 |
+
|
| 73 |
+
0. Definitions.
|
| 74 |
+
|
| 75 |
+
"This License" refers to version 3 of the GNU General Public License.
|
| 76 |
+
|
| 77 |
+
"Copyright" also means copyright-like laws that apply to other kinds of
|
| 78 |
+
works, such as semiconductor masks.
|
| 79 |
+
|
| 80 |
+
"The Program" refers to any copyrightable work licensed under this
|
| 81 |
+
License. Each licensee is addressed as "you". "Licensees" and
|
| 82 |
+
"recipients" may be individuals or organizations.
|
| 83 |
+
|
| 84 |
+
To "modify" a work means to copy from or adapt all or part of the work
|
| 85 |
+
in a fashion requiring copyright permission, other than the making of an
|
| 86 |
+
exact copy. The resulting work is called a "modified version" of the
|
| 87 |
+
earlier work or a work "based on" the earlier work.
|
| 88 |
+
|
| 89 |
+
A "covered work" means either the unmodified Program or a work based
|
| 90 |
+
on the Program.
|
| 91 |
+
|
| 92 |
+
To "propagate" a work means to do anything with it that, without
|
| 93 |
+
permission, would make you directly or secondarily liable for
|
| 94 |
+
infringement under applicable copyright law, except executing it on a
|
| 95 |
+
computer or modifying a private copy. Propagation includes copying,
|
| 96 |
+
distribution (with or without modification), making available to the
|
| 97 |
+
public, and in some countries other activities as well.
|
| 98 |
+
|
| 99 |
+
To "convey" a work means any kind of propagation that enables other
|
| 100 |
+
parties to make or receive copies. Mere interaction with a user through
|
| 101 |
+
a computer network, with no transfer of a copy, is not conveying.
|
| 102 |
+
|
| 103 |
+
An interactive user interface displays "Appropriate Legal Notices"
|
| 104 |
+
to the extent that it includes a convenient and prominently visible
|
| 105 |
+
feature that (1) displays an appropriate copyright notice, and (2)
|
| 106 |
+
tells the user that there is no warranty for the work (except to the
|
| 107 |
+
extent that warranties are provided), that licensees may convey the
|
| 108 |
+
work under this License, and how to view a copy of this License. If
|
| 109 |
+
the interface presents a list of user commands or options, such as a
|
| 110 |
+
menu, a prominent item in the list meets this criterion.
|
| 111 |
+
|
| 112 |
+
1. Source Code.
|
| 113 |
+
|
| 114 |
+
The "source code" for a work means the preferred form of the work
|
| 115 |
+
for making modifications to it. "Object code" means any non-source
|
| 116 |
+
form of a work.
|
| 117 |
+
|
| 118 |
+
A "Standard Interface" means an interface that either is an official
|
| 119 |
+
standard defined by a recognized standards body, or, in the case of
|
| 120 |
+
interfaces specified for a particular programming language, one that
|
| 121 |
+
is widely used among developers working in that language.
|
| 122 |
+
|
| 123 |
+
The "System Libraries" of an executable work include anything, other
|
| 124 |
+
than the work as a whole, that (a) is included in the normal form of
|
| 125 |
+
packaging a Major Component, but which is not part of that Major
|
| 126 |
+
Component, and (b) serves only to enable use of the work with that
|
| 127 |
+
Major Component, or to implement a Standard Interface for which an
|
| 128 |
+
implementation is available to the public in source code form. A
|
| 129 |
+
"Major Component", in this context, means a major essential component
|
| 130 |
+
(kernel, window system, and so on) of the specific operating system
|
| 131 |
+
(if any) on which the executable work runs, or a compiler used to
|
| 132 |
+
produce the work, or an object code interpreter used to run it.
|
| 133 |
+
|
| 134 |
+
The "Corresponding Source" for a work in object code form means all
|
| 135 |
+
the source code needed to generate, install, and (for an executable
|
| 136 |
+
work) run the object code and to modify the work, including scripts to
|
| 137 |
+
control those activities. However, it does not include the work's
|
| 138 |
+
System Libraries, or general-purpose tools or generally available free
|
| 139 |
+
programs which are used unmodified in performing those activities but
|
| 140 |
+
which are not part of the work. For example, Corresponding Source
|
| 141 |
+
includes interface definition files associated with source files for
|
| 142 |
+
the work, and the source code for shared libraries and dynamically
|
| 143 |
+
linked subprograms that the work is specifically designed to require,
|
| 144 |
+
such as by intimate data communication or control flow between those
|
| 145 |
+
subprograms and other parts of the work.
|
| 146 |
+
|
| 147 |
+
The Corresponding Source need not include anything that users
|
| 148 |
+
can regenerate automatically from other parts of the Corresponding
|
| 149 |
+
Source.
|
| 150 |
+
|
| 151 |
+
The Corresponding Source for a work in source code form is that
|
| 152 |
+
same work.
|
| 153 |
+
|
| 154 |
+
2. Basic Permissions.
|
| 155 |
+
|
| 156 |
+
All rights granted under this License are granted for the term of
|
| 157 |
+
copyright on the Program, and are irrevocable provided the stated
|
| 158 |
+
conditions are met. This License explicitly affirms your unlimited
|
| 159 |
+
permission to run the unmodified Program. The output from running a
|
| 160 |
+
covered work is covered by this License only if the output, given its
|
| 161 |
+
content, constitutes a covered work. This License acknowledges your
|
| 162 |
+
rights of fair use or other equivalent, as provided by copyright law.
|
| 163 |
+
|
| 164 |
+
You may make, run and propagate covered works that you do not
|
| 165 |
+
convey, without conditions so long as your license otherwise remains
|
| 166 |
+
in force. You may convey covered works to others for the sole purpose
|
| 167 |
+
of having them make modifications exclusively for you, or provide you
|
| 168 |
+
with facilities for running those works, provided that you comply with
|
| 169 |
+
the terms of this License in conveying all material for which you do
|
| 170 |
+
not control copyright. Those thus making or running the covered works
|
| 171 |
+
for you must do so exclusively on your behalf, under your direction
|
| 172 |
+
and control, on terms that prohibit them from making any copies of
|
| 173 |
+
your copyrighted material outside their relationship with you.
|
| 174 |
+
|
| 175 |
+
Conveying under any other circumstances is permitted solely under
|
| 176 |
+
the conditions stated below. Sublicensing is not allowed; section 10
|
| 177 |
+
makes it unnecessary.
|
| 178 |
+
|
| 179 |
+
3. Protecting Users' Legal Rights From Anti-Circumvention Law.
|
| 180 |
+
|
| 181 |
+
No covered work shall be deemed part of an effective technological
|
| 182 |
+
measure under any applicable law fulfilling obligations under article
|
| 183 |
+
11 of the WIPO copyright treaty adopted on 20 December 1996, or
|
| 184 |
+
similar laws prohibiting or restricting circumvention of such
|
| 185 |
+
measures.
|
| 186 |
+
|
| 187 |
+
When you convey a covered work, you waive any legal power to forbid
|
| 188 |
+
circumvention of technological measures to the extent such circumvention
|
| 189 |
+
is effected by exercising rights under this License with respect to
|
| 190 |
+
the covered work, and you disclaim any intention to limit operation or
|
| 191 |
+
modification of the work as a means of enforcing, against the work's
|
| 192 |
+
users, your or third parties' legal rights to forbid circumvention of
|
| 193 |
+
technological measures.
|
| 194 |
+
|
| 195 |
+
4. Conveying Verbatim Copies.
|
| 196 |
+
|
| 197 |
+
You may convey verbatim copies of the Program's source code as you
|
| 198 |
+
receive it, in any medium, provided that you conspicuously and
|
| 199 |
+
appropriately publish on each copy an appropriate copyright notice;
|
| 200 |
+
keep intact all notices stating that this License and any
|
| 201 |
+
non-permissive terms added in accord with section 7 apply to the code;
|
| 202 |
+
keep intact all notices of the absence of any warranty; and give all
|
| 203 |
+
recipients a copy of this License along with the Program.
|
| 204 |
+
|
| 205 |
+
You may charge any price or no price for each copy that you convey,
|
| 206 |
+
and you may offer support or warranty protection for a fee.
|
| 207 |
+
|
| 208 |
+
5. Conveying Modified Source Versions.
|
| 209 |
+
|
| 210 |
+
You may convey a work based on the Program, or the modifications to
|
| 211 |
+
produce it from the Program, in the form of source code under the
|
| 212 |
+
terms of section 4, provided that you also meet all of these conditions:
|
| 213 |
+
|
| 214 |
+
a) The work must carry prominent notices stating that you modified
|
| 215 |
+
it, and giving a relevant date.
|
| 216 |
+
|
| 217 |
+
b) The work must carry prominent notices stating that it is
|
| 218 |
+
released under this License and any conditions added under section
|
| 219 |
+
7. This requirement modifies the requirement in section 4 to
|
| 220 |
+
"keep intact all notices".
|
| 221 |
+
|
| 222 |
+
c) You must license the entire work, as a whole, under this
|
| 223 |
+
License to anyone who comes into possession of a copy. This
|
| 224 |
+
License will therefore apply, along with any applicable section 7
|
| 225 |
+
additional terms, to the whole of the work, and all its parts,
|
| 226 |
+
regardless of how they are packaged. This License gives no
|
| 227 |
+
permission to license the work in any other way, but it does not
|
| 228 |
+
invalidate such permission if you have separately received it.
|
| 229 |
+
|
| 230 |
+
d) If the work has interactive user interfaces, each must display
|
| 231 |
+
Appropriate Legal Notices; however, if the Program has interactive
|
| 232 |
+
interfaces that do not display Appropriate Legal Notices, your
|
| 233 |
+
work need not make them do so.
|
| 234 |
+
|
| 235 |
+
A compilation of a covered work with other separate and independent
|
| 236 |
+
works, which are not by their nature extensions of the covered work,
|
| 237 |
+
and which are not combined with it such as to form a larger program,
|
| 238 |
+
in or on a volume of a storage or distribution medium, is called an
|
| 239 |
+
"aggregate" if the compilation and its resulting copyright are not
|
| 240 |
+
used to limit the access or legal rights of the compilation's users
|
| 241 |
+
beyond what the individual works permit. Inclusion of a covered work
|
| 242 |
+
in an aggregate does not cause this License to apply to the other
|
| 243 |
+
parts of the aggregate.
|
| 244 |
+
|
| 245 |
+
6. Conveying Non-Source Forms.
|
| 246 |
+
|
| 247 |
+
You may convey a covered work in object code form under the terms
|
| 248 |
+
of sections 4 and 5, provided that you also convey the
|
| 249 |
+
machine-readable Corresponding Source under the terms of this License,
|
| 250 |
+
in one of these ways:
|
| 251 |
+
|
| 252 |
+
a) Convey the object code in, or embodied in, a physical product
|
| 253 |
+
(including a physical distribution medium), accompanied by the
|
| 254 |
+
Corresponding Source fixed on a durable physical medium
|
| 255 |
+
customarily used for software interchange.
|
| 256 |
+
|
| 257 |
+
b) Convey the object code in, or embodied in, a physical product
|
| 258 |
+
(including a physical distribution medium), accompanied by a
|
| 259 |
+
written offer, valid for at least three years and valid for as
|
| 260 |
+
long as you offer spare parts or customer support for that product
|
| 261 |
+
model, to give anyone who possesses the object code either (1) a
|
| 262 |
+
copy of the Corresponding Source for all the software in the
|
| 263 |
+
product that is covered by this License, on a durable physical
|
| 264 |
+
medium customarily used for software interchange, for a price no
|
| 265 |
+
more than your reasonable cost of physically performing this
|
| 266 |
+
conveying of source, or (2) access to copy the
|
| 267 |
+
Corresponding Source from a network server at no charge.
|
| 268 |
+
|
| 269 |
+
c) Convey individual copies of the object code with a copy of the
|
| 270 |
+
written offer to provide the Corresponding Source. This
|
| 271 |
+
alternative is allowed only occasionally and noncommercially, and
|
| 272 |
+
only if you received the object code with such an offer, in accord
|
| 273 |
+
with subsection 6b.
|
| 274 |
+
|
| 275 |
+
d) Convey the object code by offering access from a designated
|
| 276 |
+
place (gratis or for a charge), and offer equivalent access to the
|
| 277 |
+
Corresponding Source in the same way through the same place at no
|
| 278 |
+
further charge. You need not require recipients to copy the
|
| 279 |
+
Corresponding Source along with the object code. If the place to
|
| 280 |
+
copy the object code is a network server, the Corresponding Source
|
| 281 |
+
may be on a different server (operated by you or a third party)
|
| 282 |
+
that supports equivalent copying facilities, provided you maintain
|
| 283 |
+
clear directions next to the object code saying where to find the
|
| 284 |
+
Corresponding Source. Regardless of what server hosts the
|
| 285 |
+
Corresponding Source, you remain obligated to ensure that it is
|
| 286 |
+
available for as long as needed to satisfy these requirements.
|
| 287 |
+
|
| 288 |
+
e) Convey the object code using peer-to-peer transmission, provided
|
| 289 |
+
you inform other peers where the object code and Corresponding
|
| 290 |
+
Source of the work are being offered to the general public at no
|
| 291 |
+
charge under subsection 6d.
|
| 292 |
+
|
| 293 |
+
A separable portion of the object code, whose source code is excluded
|
| 294 |
+
from the Corresponding Source as a System Library, need not be
|
| 295 |
+
included in conveying the object code work.
|
| 296 |
+
|
| 297 |
+
A "User Product" is either (1) a "consumer product", which means any
|
| 298 |
+
tangible personal property which is normally used for personal, family,
|
| 299 |
+
or household purposes, or (2) anything designed or sold for incorporation
|
| 300 |
+
into a dwelling. In determining whether a product is a consumer product,
|
| 301 |
+
doubtful cases shall be resolved in favor of coverage. For a particular
|
| 302 |
+
product received by a particular user, "normally used" refers to a
|
| 303 |
+
typical or common use of that class of product, regardless of the status
|
| 304 |
+
of the particular user or of the way in which the particular user
|
| 305 |
+
actually uses, or expects or is expected to use, the product. A product
|
| 306 |
+
is a consumer product regardless of whether the product has substantial
|
| 307 |
+
commercial, industrial or non-consumer uses, unless such uses represent
|
| 308 |
+
the only significant mode of use of the product.
|
| 309 |
+
|
| 310 |
+
"Installation Information" for a User Product means any methods,
|
| 311 |
+
procedures, authorization keys, or other information required to install
|
| 312 |
+
and execute modified versions of a covered work in that User Product from
|
| 313 |
+
a modified version of its Corresponding Source. The information must
|
| 314 |
+
suffice to ensure that the continued functioning of the modified object
|
| 315 |
+
code is in no case prevented or interfered with solely because
|
| 316 |
+
modification has been made.
|
| 317 |
+
|
| 318 |
+
If you convey an object code work under this section in, or with, or
|
| 319 |
+
specifically for use in, a User Product, and the conveying occurs as
|
| 320 |
+
part of a transaction in which the right of possession and use of the
|
| 321 |
+
User Product is transferred to the recipient in perpetuity or for a
|
| 322 |
+
fixed term (regardless of how the transaction is characterized), the
|
| 323 |
+
Corresponding Source conveyed under this section must be accompanied
|
| 324 |
+
by the Installation Information. But this requirement does not apply
|
| 325 |
+
if neither you nor any third party retains the ability to install
|
| 326 |
+
modified object code on the User Product (for example, the work has
|
| 327 |
+
been installed in ROM).
|
| 328 |
+
|
| 329 |
+
The requirement to provide Installation Information does not include a
|
| 330 |
+
requirement to continue to provide support service, warranty, or updates
|
| 331 |
+
for a work that has been modified or installed by the recipient, or for
|
| 332 |
+
the User Product in which it has been modified or installed. Access to a
|
| 333 |
+
network may be denied when the modification itself materially and
|
| 334 |
+
adversely affects the operation of the network or violates the rules and
|
| 335 |
+
protocols for communication across the network.
|
| 336 |
+
|
| 337 |
+
Corresponding Source conveyed, and Installation Information provided,
|
| 338 |
+
in accord with this section must be in a format that is publicly
|
| 339 |
+
documented (and with an implementation available to the public in
|
| 340 |
+
source code form), and must require no special password or key for
|
| 341 |
+
unpacking, reading or copying.
|
| 342 |
+
|
| 343 |
+
7. Additional Terms.
|
| 344 |
+
|
| 345 |
+
"Additional permissions" are terms that supplement the terms of this
|
| 346 |
+
License by making exceptions from one or more of its conditions.
|
| 347 |
+
Additional permissions that are applicable to the entire Program shall
|
| 348 |
+
be treated as though they were included in this License, to the extent
|
| 349 |
+
that they are valid under applicable law. If additional permissions
|
| 350 |
+
apply only to part of the Program, that part may be used separately
|
| 351 |
+
under those permissions, but the entire Program remains governed by
|
| 352 |
+
this License without regard to the additional permissions.
|
| 353 |
+
|
| 354 |
+
When you convey a copy of a covered work, you may at your option
|
| 355 |
+
remove any additional permissions from that copy, or from any part of
|
| 356 |
+
it. (Additional permissions may be written to require their own
|
| 357 |
+
removal in certain cases when you modify the work.) You may place
|
| 358 |
+
additional permissions on material, added by you to a covered work,
|
| 359 |
+
for which you have or can give appropriate copyright permission.
|
| 360 |
+
|
| 361 |
+
Notwithstanding any other provision of this License, for material you
|
| 362 |
+
add to a covered work, you may (if authorized by the copyright holders of
|
| 363 |
+
that material) supplement the terms of this License with terms:
|
| 364 |
+
|
| 365 |
+
a) Disclaiming warranty or limiting liability differently from the
|
| 366 |
+
terms of sections 15 and 16 of this License; or
|
| 367 |
+
|
| 368 |
+
b) Requiring preservation of specified reasonable legal notices or
|
| 369 |
+
author attributions in that material or in the Appropriate Legal
|
| 370 |
+
Notices displayed by works containing it; or
|
| 371 |
+
|
| 372 |
+
c) Prohibiting misrepresentation of the origin of that material, or
|
| 373 |
+
requiring that modified versions of such material be marked in
|
| 374 |
+
reasonable ways as different from the original version; or
|
| 375 |
+
|
| 376 |
+
d) Limiting the use for publicity purposes of names of licensors or
|
| 377 |
+
authors of the material; or
|
| 378 |
+
|
| 379 |
+
e) Declining to grant rights under trademark law for use of some
|
| 380 |
+
trade names, trademarks, or service marks; or
|
| 381 |
+
|
| 382 |
+
f) Requiring indemnification of licensors and authors of that
|
| 383 |
+
material by anyone who conveys the material (or modified versions of
|
| 384 |
+
it) with contractual assumptions of liability to the recipient, for
|
| 385 |
+
any liability that these contractual assumptions directly impose on
|
| 386 |
+
those licensors and authors.
|
| 387 |
+
|
| 388 |
+
All other non-permissive additional terms are considered "further
|
| 389 |
+
restrictions" within the meaning of section 10. If the Program as you
|
| 390 |
+
received it, or any part of it, contains a notice stating that it is
|
| 391 |
+
governed by this License along with a term that is a further
|
| 392 |
+
restriction, you may remove that term. If a license document contains
|
| 393 |
+
a further restriction but permits relicensing or conveying under this
|
| 394 |
+
License, you may add to a covered work material governed by the terms
|
| 395 |
+
of that license document, provided that the further restriction does
|
| 396 |
+
not survive such relicensing or conveying.
|
| 397 |
+
|
| 398 |
+
If you add terms to a covered work in accord with this section, you
|
| 399 |
+
must place, in the relevant source files, a statement of the
|
| 400 |
+
additional terms that apply to those files, or a notice indicating
|
| 401 |
+
where to find the applicable terms.
|
| 402 |
+
|
| 403 |
+
Additional terms, permissive or non-permissive, may be stated in the
|
| 404 |
+
form of a separately written license, or stated as exceptions;
|
| 405 |
+
the above requirements apply either way.
|
| 406 |
+
|
| 407 |
+
8. Termination.
|
| 408 |
+
|
| 409 |
+
You may not propagate or modify a covered work except as expressly
|
| 410 |
+
provided under this License. Any attempt otherwise to propagate or
|
| 411 |
+
modify it is void, and will automatically terminate your rights under
|
| 412 |
+
this License (including any patent licenses granted under the third
|
| 413 |
+
paragraph of section 11).
|
| 414 |
+
|
| 415 |
+
However, if you cease all violation of this License, then your
|
| 416 |
+
license from a particular copyright holder is reinstated (a)
|
| 417 |
+
provisionally, unless and until the copyright holder explicitly and
|
| 418 |
+
finally terminates your license, and (b) permanently, if the copyright
|
| 419 |
+
holder fails to notify you of the violation by some reasonable means
|
| 420 |
+
prior to 60 days after the cessation.
|
| 421 |
+
|
| 422 |
+
Moreover, your license from a particular copyright holder is
|
| 423 |
+
reinstated permanently if the copyright holder notifies you of the
|
| 424 |
+
violation by some reasonable means, this is the first time you have
|
| 425 |
+
received notice of violation of this License (for any work) from that
|
| 426 |
+
copyright holder, and you cure the violation prior to 30 days after
|
| 427 |
+
your receipt of the notice.
|
| 428 |
+
|
| 429 |
+
Termination of your rights under this section does not terminate the
|
| 430 |
+
licenses of parties who have received copies or rights from you under
|
| 431 |
+
this License. If your rights have been terminated and not permanently
|
| 432 |
+
reinstated, you do not qualify to receive new licenses for the same
|
| 433 |
+
material under section 10.
|
| 434 |
+
|
| 435 |
+
9. Acceptance Not Required for Having Copies.
|
| 436 |
+
|
| 437 |
+
You are not required to accept this License in order to receive or
|
| 438 |
+
run a copy of the Program. Ancillary propagation of a covered work
|
| 439 |
+
occurring solely as a consequence of using peer-to-peer transmission
|
| 440 |
+
to receive a copy likewise does not require acceptance. However,
|
| 441 |
+
nothing other than this License grants you permission to propagate or
|
| 442 |
+
modify any covered work. These actions infringe copyright if you do
|
| 443 |
+
not accept this License. Therefore, by modifying or propagating a
|
| 444 |
+
covered work, you indicate your acceptance of this License to do so.
|
| 445 |
+
|
| 446 |
+
10. Automatic Licensing of Downstream Recipients.
|
| 447 |
+
|
| 448 |
+
Each time you convey a covered work, the recipient automatically
|
| 449 |
+
receives a license from the original licensors, to run, modify and
|
| 450 |
+
propagate that work, subject to this License. You are not responsible
|
| 451 |
+
for enforcing compliance by third parties with this License.
|
| 452 |
+
|
| 453 |
+
An "entity transaction" is a transaction transferring control of an
|
| 454 |
+
organization, or substantially all assets of one, or subdividing an
|
| 455 |
+
organization, or merging organizations. If propagation of a covered
|
| 456 |
+
work results from an entity transaction, each party to that
|
| 457 |
+
transaction who receives a copy of the work also receives whatever
|
| 458 |
+
licenses to the work the party's predecessor in interest had or could
|
| 459 |
+
give under the previous paragraph, plus a right to possession of the
|
| 460 |
+
Corresponding Source of the work from the predecessor in interest, if
|
| 461 |
+
the predecessor has it or can get it with reasonable efforts.
|
| 462 |
+
|
| 463 |
+
You may not impose any further restrictions on the exercise of the
|
| 464 |
+
rights granted or affirmed under this License. For example, you may
|
| 465 |
+
not impose a license fee, royalty, or other charge for exercise of
|
| 466 |
+
rights granted under this License, and you may not initiate litigation
|
| 467 |
+
(including a cross-claim or counterclaim in a lawsuit) alleging that
|
| 468 |
+
any patent claim is infringed by making, using, selling, offering for
|
| 469 |
+
sale, or importing the Program or any portion of it.
|
| 470 |
+
|
| 471 |
+
11. Patents.
|
| 472 |
+
|
| 473 |
+
A "contributor" is a copyright holder who authorizes use under this
|
| 474 |
+
License of the Program or a work on which the Program is based. The
|
| 475 |
+
work thus licensed is called the contributor's "contributor version".
|
| 476 |
+
|
| 477 |
+
A contributor's "essential patent claims" are all patent claims
|
| 478 |
+
owned or controlled by the contributor, whether already acquired or
|
| 479 |
+
hereafter acquired, that would be infringed by some manner, permitted
|
| 480 |
+
by this License, of making, using, or selling its contributor version,
|
| 481 |
+
but do not include claims that would be infringed only as a
|
| 482 |
+
consequence of further modification of the contributor version. For
|
| 483 |
+
purposes of this definition, "control" includes the right to grant
|
| 484 |
+
patent sublicenses in a manner consistent with the requirements of
|
| 485 |
+
this License.
|
| 486 |
+
|
| 487 |
+
Each contributor grants you a non-exclusive, worldwide, royalty-free
|
| 488 |
+
patent license under the contributor's essential patent claims, to
|
| 489 |
+
make, use, sell, offer for sale, import and otherwise run, modify and
|
| 490 |
+
propagate the contents of its contributor version.
|
| 491 |
+
|
| 492 |
+
In the following three paragraphs, a "patent license" is any express
|
| 493 |
+
agreement or commitment, however denominated, not to enforce a patent
|
| 494 |
+
(such as an express permission to practice a patent or covenant not to
|
| 495 |
+
sue for patent infringement). To "grant" such a patent license to a
|
| 496 |
+
party means to make such an agreement or commitment not to enforce a
|
| 497 |
+
patent against the party.
|
| 498 |
+
|
| 499 |
+
If you convey a covered work, knowingly relying on a patent license,
|
| 500 |
+
and the Corresponding Source of the work is not available for anyone
|
| 501 |
+
to copy, free of charge and under the terms of this License, through a
|
| 502 |
+
publicly available network server or other readily accessible means,
|
| 503 |
+
then you must either (1) cause the Corresponding Source to be so
|
| 504 |
+
available, or (2) arrange to deprive yourself of the benefit of the
|
| 505 |
+
patent license for this particular work, or (3) arrange, in a manner
|
| 506 |
+
consistent with the requirements of this License, to extend the patent
|
| 507 |
+
license to downstream recipients. "Knowingly relying" means you have
|
| 508 |
+
actual knowledge that, but for the patent license, your conveying the
|
| 509 |
+
covered work in a country, or your recipient's use of the covered work
|
| 510 |
+
in a country, would infringe one or more identifiable patents in that
|
| 511 |
+
country that you have reason to believe are valid.
|
| 512 |
+
|
| 513 |
+
If, pursuant to or in connection with a single transaction or
|
| 514 |
+
arrangement, you convey, or propagate by procuring conveyance of, a
|
| 515 |
+
covered work, and grant a patent license to some of the parties
|
| 516 |
+
receiving the covered work authorizing them to use, propagate, modify
|
| 517 |
+
or convey a specific copy of the covered work, then the patent license
|
| 518 |
+
you grant is automatically extended to all recipients of the covered
|
| 519 |
+
work and works based on it.
|
| 520 |
+
|
| 521 |
+
A patent license is "discriminatory" if it does not include within
|
| 522 |
+
the scope of its coverage, prohibits the exercise of, or is
|
| 523 |
+
conditioned on the non-exercise of one or more of the rights that are
|
| 524 |
+
specifically granted under this License. You may not convey a covered
|
| 525 |
+
work if you are a party to an arrangement with a third party that is
|
| 526 |
+
in the business of distributing software, under which you make payment
|
| 527 |
+
to the third party based on the extent of your activity of conveying
|
| 528 |
+
the work, and under which the third party grants, to any of the
|
| 529 |
+
parties who would receive the covered work from you, a discriminatory
|
| 530 |
+
patent license (a) in connection with copies of the covered work
|
| 531 |
+
conveyed by you (or copies made from those copies), or (b) primarily
|
| 532 |
+
for and in connection with specific products or compilations that
|
| 533 |
+
contain the covered work, unless you entered into that arrangement,
|
| 534 |
+
or that patent license was granted, prior to 28 March 2007.
|
| 535 |
+
|
| 536 |
+
Nothing in this License shall be construed as excluding or limiting
|
| 537 |
+
any implied license or other defenses to infringement that may
|
| 538 |
+
otherwise be available to you under applicable patent law.
|
| 539 |
+
|
| 540 |
+
12. No Surrender of Others' Freedom.
|
| 541 |
+
|
| 542 |
+
If conditions are imposed on you (whether by court order, agreement or
|
| 543 |
+
otherwise) that contradict the conditions of this License, they do not
|
| 544 |
+
excuse you from the conditions of this License. If you cannot convey a
|
| 545 |
+
covered work so as to satisfy simultaneously your obligations under this
|
| 546 |
+
License and any other pertinent obligations, then as a consequence you may
|
| 547 |
+
not convey it at all. For example, if you agree to terms that obligate you
|
| 548 |
+
to collect a royalty for further conveying from those to whom you convey
|
| 549 |
+
the Program, the only way you could satisfy both those terms and this
|
| 550 |
+
License would be to refrain entirely from conveying the Program.
|
| 551 |
+
|
| 552 |
+
13. Use with the GNU Affero General Public License.
|
| 553 |
+
|
| 554 |
+
Notwithstanding any other provision of this License, you have
|
| 555 |
+
permission to link or combine any covered work with a work licensed
|
| 556 |
+
under version 3 of the GNU Affero General Public License into a single
|
| 557 |
+
combined work, and to convey the resulting work. The terms of this
|
| 558 |
+
License will continue to apply to the part which is the covered work,
|
| 559 |
+
but the special requirements of the GNU Affero General Public License,
|
| 560 |
+
section 13, concerning interaction through a network will apply to the
|
| 561 |
+
combination as such.
|
| 562 |
+
|
| 563 |
+
14. Revised Versions of this License.
|
| 564 |
+
|
| 565 |
+
The Free Software Foundation may publish revised and/or new versions of
|
| 566 |
+
the GNU General Public License from time to time. Such new versions will
|
| 567 |
+
be similar in spirit to the present version, but may differ in detail to
|
| 568 |
+
address new problems or concerns.
|
| 569 |
+
|
| 570 |
+
Each version is given a distinguishing version number. If the
|
| 571 |
+
Program specifies that a certain numbered version of the GNU General
|
| 572 |
+
Public License "or any later version" applies to it, you have the
|
| 573 |
+
option of following the terms and conditions either of that numbered
|
| 574 |
+
version or of any later version published by the Free Software
|
| 575 |
+
Foundation. If the Program does not specify a version number of the
|
| 576 |
+
GNU General Public License, you may choose any version ever published
|
| 577 |
+
by the Free Software Foundation.
|
| 578 |
+
|
| 579 |
+
If the Program specifies that a proxy can decide which future
|
| 580 |
+
versions of the GNU General Public License can be used, that proxy's
|
| 581 |
+
public statement of acceptance of a version permanently authorizes you
|
| 582 |
+
to choose that version for the Program.
|
| 583 |
+
|
| 584 |
+
Later license versions may give you additional or different
|
| 585 |
+
permissions. However, no additional obligations are imposed on any
|
| 586 |
+
author or copyright holder as a result of your choosing to follow a
|
| 587 |
+
later version.
|
| 588 |
+
|
| 589 |
+
15. Disclaimer of Warranty.
|
| 590 |
+
|
| 591 |
+
THERE IS NO WARRANTY FOR THE PROGRAM, TO THE EXTENT PERMITTED BY
|
| 592 |
+
APPLICABLE LAW. EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT
|
| 593 |
+
HOLDERS AND/OR OTHER PARTIES PROVIDE THE PROGRAM "AS IS" WITHOUT WARRANTY
|
| 594 |
+
OF ANY KIND, EITHER EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO,
|
| 595 |
+
THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
|
| 596 |
+
PURPOSE. THE ENTIRE RISK AS TO THE QUALITY AND PERFORMANCE OF THE PROGRAM
|
| 597 |
+
IS WITH YOU. SHOULD THE PROGRAM PROVE DEFECTIVE, YOU ASSUME THE COST OF
|
| 598 |
+
ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
|
| 599 |
+
|
| 600 |
+
16. Limitation of Liability.
|
| 601 |
+
|
| 602 |
+
IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING
|
| 603 |
+
WILL ANY COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MODIFIES AND/OR CONVEYS
|
| 604 |
+
THE PROGRAM AS PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY
|
| 605 |
+
GENERAL, SPECIAL, INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE
|
| 606 |
+
USE OR INABILITY TO USE THE PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF
|
| 607 |
+
DATA OR DATA BEING RENDERED INACCURATE OR LOSSES SUSTAINED BY YOU OR THIRD
|
| 608 |
+
PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE WITH ANY OTHER PROGRAMS),
|
| 609 |
+
EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE POSSIBILITY OF
|
| 610 |
+
SUCH DAMAGES.
|
| 611 |
+
|
| 612 |
+
17. Interpretation of Sections 15 and 16.
|
| 613 |
+
|
| 614 |
+
If the disclaimer of warranty and limitation of liability provided
|
| 615 |
+
above cannot be given local legal effect according to their terms,
|
| 616 |
+
reviewing courts shall apply local law that most closely approximates
|
| 617 |
+
an absolute waiver of all civil liability in connection with the
|
| 618 |
+
Program, unless a warranty or assumption of liability accompanies a
|
| 619 |
+
copy of the Program in return for a fee.
|
| 620 |
+
|
| 621 |
+
END OF TERMS AND CONDITIONS
|
| 622 |
+
|
| 623 |
+
How to Apply These Terms to Your New Programs
|
| 624 |
+
|
| 625 |
+
If you develop a new program, and you want it to be of the greatest
|
| 626 |
+
possible use to the public, the best way to achieve this is to make it
|
| 627 |
+
free software which everyone can redistribute and change under these terms.
|
| 628 |
+
|
| 629 |
+
To do so, attach the following notices to the program. It is safest
|
| 630 |
+
to attach them to the start of each source file to most effectively
|
| 631 |
+
state the exclusion of warranty; and each file should have at least
|
| 632 |
+
the "copyright" line and a pointer to where the full notice is found.
|
| 633 |
+
|
| 634 |
+
{one line to give the program's name and a brief idea of what it does.}
|
| 635 |
+
Copyright (C) {year} {name of author}
|
| 636 |
+
|
| 637 |
+
This program is free software: you can redistribute it and/or modify
|
| 638 |
+
it under the terms of the GNU General Public License as published by
|
| 639 |
+
the Free Software Foundation, either version 3 of the License, or
|
| 640 |
+
(at your option) any later version.
|
| 641 |
+
|
| 642 |
+
This program is distributed in the hope that it will be useful,
|
| 643 |
+
but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 644 |
+
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 645 |
+
GNU General Public License for more details.
|
| 646 |
+
|
| 647 |
+
You should have received a copy of the GNU General Public License
|
| 648 |
+
along with this program. If not, see <http://www.gnu.org/licenses/>.
|
| 649 |
+
|
| 650 |
+
Also add information on how to contact you by electronic and paper mail.
|
| 651 |
+
|
| 652 |
+
If the program does terminal interaction, make it output a short
|
| 653 |
+
notice like this when it starts in an interactive mode:
|
| 654 |
+
|
| 655 |
+
{project} Copyright (C) {year} {fullname}
|
| 656 |
+
This program comes with ABSOLUTELY NO WARRANTY; for details type `show w'.
|
| 657 |
+
This is free software, and you are welcome to redistribute it
|
| 658 |
+
under certain conditions; type `show c' for details.
|
| 659 |
+
|
| 660 |
+
The hypothetical commands `show w' and `show c' should show the appropriate
|
| 661 |
+
parts of the General Public License. Of course, your program's commands
|
| 662 |
+
might be different; for a GUI interface, you would use an "about box".
|
| 663 |
+
|
| 664 |
+
You should also get your employer (if you work as a programmer) or school,
|
| 665 |
+
if any, to sign a "copyright disclaimer" for the program, if necessary.
|
| 666 |
+
For more information on this, and how to apply and follow the GNU GPL, see
|
| 667 |
+
<http://www.gnu.org/licenses/>.
|
| 668 |
+
|
| 669 |
+
The GNU General Public License does not permit incorporating your program
|
| 670 |
+
into proprietary programs. If your program is a subroutine library, you
|
| 671 |
+
may consider it more useful to permit linking proprietary applications with
|
| 672 |
+
the library. If this is what you want to do, use the GNU Lesser General
|
| 673 |
+
Public License instead of this License. But first, please read
|
| 674 |
+
<http://www.gnu.org/philosophy/why-not-lgpl.html>.
|
MANIFEST.in
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
include b3clf/pre_trained/*
|
| 2 |
+
include b3clf/feature_list.txt
|
| 3 |
+
include b3clf/data/B3clf_thresholds.xlsx
|
b3clf/BBB_general_workflow_v4.png
ADDED

|
b3clf/__init__.py
ADDED
|
@@ -0,0 +1,31 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""Package for BBB predictions."""
|
| 25 |
+
|
| 26 |
+
try:
|
| 27 |
+
from .version import __version__
|
| 28 |
+
except ImportError:
|
| 29 |
+
__version__ = "0.0.0.post0"
|
| 30 |
+
|
| 31 |
+
from .b3clf import b3clf
|
b3clf/__main__.py
ADDED
|
@@ -0,0 +1,109 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""Package for BBB predictions."""
|
| 25 |
+
import argparse
|
| 26 |
+
|
| 27 |
+
from .b3clf import b3clf
|
| 28 |
+
|
| 29 |
+
try:
|
| 30 |
+
from .version import __version__
|
| 31 |
+
except ImportError:
|
| 32 |
+
__version__ = "0.0.0.post0"
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
def main():
|
| 36 |
+
# https://docs.python.org/3/library/argparse.html
|
| 37 |
+
parser = argparse.ArgumentParser(
|
| 38 |
+
description="b3clf predicts if molecules can pass blood-brain barrier with resampling "
|
| 39 |
+
"strategies.",
|
| 40 |
+
)
|
| 41 |
+
parser.add_argument("-mol",
|
| 42 |
+
default="input.sdf",
|
| 43 |
+
type=str,
|
| 44 |
+
help="Input file with descriptors.")
|
| 45 |
+
parser.add_argument("-sep",
|
| 46 |
+
type=str,
|
| 47 |
+
default="\s+|\t+",
|
| 48 |
+
help="""Separator for input file. Default="\s+|\\t+".""")
|
| 49 |
+
parser.add_argument("-clf",
|
| 50 |
+
type=str,
|
| 51 |
+
default="xgb",
|
| 52 |
+
help="Classification algorithm type. Default=xgb.")
|
| 53 |
+
parser.add_argument("-sampling",
|
| 54 |
+
type=str,
|
| 55 |
+
default="classic_ADASYN",
|
| 56 |
+
help="Resampling method type. Default=classic_ADASYN.")
|
| 57 |
+
parser.add_argument("-output",
|
| 58 |
+
type=str,
|
| 59 |
+
default="B3clf_output.xlsx",
|
| 60 |
+
help="Name of output file, CSV or XLSX format. Default=B3clf_output.xlsx.")
|
| 61 |
+
parser.add_argument("-verbose",
|
| 62 |
+
type=int,
|
| 63 |
+
default=1,
|
| 64 |
+
help="If verbose is not zero, B3clf will print out the predictions. "
|
| 65 |
+
"Default=1.")
|
| 66 |
+
parser.add_argument("-random_seed",
|
| 67 |
+
type=int,
|
| 68 |
+
default=42,
|
| 69 |
+
help="""Romdom seed to control randonness. If set to be "None", """
|
| 70 |
+
"""it will result in a randomness of the predictions. Default=42.""")
|
| 71 |
+
parser.add_argument("-time_per_mol",
|
| 72 |
+
type=int,
|
| 73 |
+
default=-1,
|
| 74 |
+
help="""Time per molecule in seconds. If set to be -1, no time limit. """
|
| 75 |
+
"""Default=-1.""")
|
| 76 |
+
parser.add_argument("-keep_features",
|
| 77 |
+
type=str,
|
| 78 |
+
default="no",
|
| 79 |
+
help="""To keep computed feature file ("yes") or not ("no"). Default=no.""")
|
| 80 |
+
parser.add_argument("-keep_sdf",
|
| 81 |
+
type=str,
|
| 82 |
+
default="no",
|
| 83 |
+
help="""To keep computed molecular geometries ("yes") or not ("no"). Default=no.""")
|
| 84 |
+
parser.add_argument("-threshold",
|
| 85 |
+
type=str,
|
| 86 |
+
default="none",
|
| 87 |
+
help="""Threshold used for the classification which can be "none", """
|
| 88 |
+
""""J_threshold" and "F_threshold". "J_threshold" will use """
|
| 89 |
+
"""threshold optimized from Youden’s J statistic. "F_threshold" will """
|
| 90 |
+
"""use threshold optimized from F score. Default="none".""")
|
| 91 |
+
args = parser.parse_args()
|
| 92 |
+
|
| 93 |
+
_ = b3clf(mol_in=args.mol,
|
| 94 |
+
sep=args.sep,
|
| 95 |
+
clf=args.clf,
|
| 96 |
+
sampling=args.sampling,
|
| 97 |
+
output=args.output,
|
| 98 |
+
verbose=args.verbose,
|
| 99 |
+
random_seed=args.random_seed,
|
| 100 |
+
time_per_mol=args.time_per_mol,
|
| 101 |
+
keep_features=args.keep_features,
|
| 102 |
+
keep_sdf=args.keep_sdf,
|
| 103 |
+
threshold=args.threshold,
|
| 104 |
+
)
|
| 105 |
+
|
| 106 |
+
|
| 107 |
+
if __name__ == "__main__":
|
| 108 |
+
"""B3clf command-line interface."""
|
| 109 |
+
main()
|
b3clf/b3clf.py
ADDED
|
@@ -0,0 +1,154 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""
|
| 25 |
+
Main B3clf Script.
|
| 26 |
+
"""
|
| 27 |
+
|
| 28 |
+
# Todo: Enable b3clf prediction without PaDeL calculation from PaDeL descriptor input
|
| 29 |
+
import os
|
| 30 |
+
|
| 31 |
+
import numpy as np
|
| 32 |
+
from .descriptor_padel import compute_descriptors
|
| 33 |
+
from .geometry_opt import geometry_optimize
|
| 34 |
+
from .utils import (get_descriptors, predict_permeability,
|
| 35 |
+
scale_descriptors, select_descriptors)
|
| 36 |
+
|
| 37 |
+
__all__ = [
|
| 38 |
+
"b3clf",
|
| 39 |
+
]
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
def b3clf(mol_in,
|
| 43 |
+
sep="\s+|\t+",
|
| 44 |
+
clf="xgb",
|
| 45 |
+
sampling="classic_ADASYN",
|
| 46 |
+
output="B3clf_output.xlsx",
|
| 47 |
+
verbose=1,
|
| 48 |
+
random_seed=42,
|
| 49 |
+
time_per_mol=-1,
|
| 50 |
+
keep_features="no",
|
| 51 |
+
keep_sdf="no",
|
| 52 |
+
threshold="none",
|
| 53 |
+
):
|
| 54 |
+
"""Use B3clf for BBB classifications with resampling strategies.
|
| 55 |
+
|
| 56 |
+
Parameters
|
| 57 |
+
----------
|
| 58 |
+
mol_in : str
|
| 59 |
+
Input molecule text fie which can be SMILES strings (file extension with .smi or .csv) or
|
| 60 |
+
SDF file format. No space is allowed for molecular name if input is a file with SMILES strings.
|
| 61 |
+
sep : str, optional
|
| 62 |
+
Separator used to parse data if a text file with SMILES strings is provided.
|
| 63 |
+
Default="\s+|\t+" which will take any space and any tab as delimiter.
|
| 64 |
+
clf: str, optional
|
| 65 |
+
Classification algorithm, which can be "dtree" for decision trees, "knn" for kNN, "logreg"
|
| 66 |
+
for logistical regression and "xgb" for XGBoost. Default="xgb".
|
| 67 |
+
sampling : str, optional
|
| 68 |
+
Sampling strategies that can be used which includes "common",
|
| 69 |
+
"RandUndersampling", "SMOTE", "borderline_SMOTE", "kmeans_SMOTE" and "classic_ADASYN". The
|
| 70 |
+
"common" denotes that no resampling strategy is employed. Default="classic_ADASYN".
|
| 71 |
+
output : str, optional
|
| 72 |
+
Output file name for the predicted results consisting molecule ID, predicted probability
|
| 73 |
+
and labels for BBB permeability.
|
| 74 |
+
verbose : int, optional
|
| 75 |
+
When verbose is zero, no results are printed out. Otherwise, the program prints the
|
| 76 |
+
predictions. Default=1.
|
| 77 |
+
random_seed : int, optional
|
| 78 |
+
Random seed for reproducibility. Default=42.
|
| 79 |
+
time_per_mol : int, optional
|
| 80 |
+
Time limit for each molecule in seconds. Default=-1, which means no time limit.
|
| 81 |
+
keep_features : str, optional
|
| 82 |
+
To keep intermediate molecular feature file, "yes" or "no". Default="no".
|
| 83 |
+
keep_sdf : str, optional
|
| 84 |
+
To keep intermediate molecular geometry file with 3D coordinates, "yes" or "no".
|
| 85 |
+
Default="no".
|
| 86 |
+
threshold : str, optional
|
| 87 |
+
To set the threshold for the predicted probability which can be "none". "J_threshold" and
|
| 88 |
+
"F_threshold". "J_threshold" will use threshold optimized from Youden’s J statistic.
|
| 89 |
+
"F_threshold" will use threshold optimized from F score. Default="none".
|
| 90 |
+
|
| 91 |
+
Returns
|
| 92 |
+
-------
|
| 93 |
+
result_df : pandas.DataFrame
|
| 94 |
+
Result of BBB predictions with molecule ID/name, predicted probability and predicted labels.
|
| 95 |
+
|
| 96 |
+
"""
|
| 97 |
+
|
| 98 |
+
# set random seed
|
| 99 |
+
if random_seed is not None:
|
| 100 |
+
rng = np.random.default_rng(random_seed)
|
| 101 |
+
|
| 102 |
+
mol_tag = os.path.basename(mol_in).split(".")[0]
|
| 103 |
+
|
| 104 |
+
features_out = f"{mol_tag}_padel_descriptors.xlsx"
|
| 105 |
+
internal_sdf = f"{mol_tag}_optimized_3d.sdf"
|
| 106 |
+
|
| 107 |
+
# Geometry optimization
|
| 108 |
+
# Input:
|
| 109 |
+
# * Either an SDF file with molecular geometries or a text file with SMILES strings
|
| 110 |
+
|
| 111 |
+
geometry_optimize(input_fname=mol_in, output_sdf=internal_sdf, sep=sep)
|
| 112 |
+
|
| 113 |
+
_ = compute_descriptors(sdf_file=internal_sdf,
|
| 114 |
+
excel_out=features_out,
|
| 115 |
+
output_csv=None,
|
| 116 |
+
timeout=None,
|
| 117 |
+
time_per_molecule=time_per_mol,
|
| 118 |
+
)
|
| 119 |
+
|
| 120 |
+
# Get computed descriptors
|
| 121 |
+
X_features, info_df = get_descriptors(df=features_out)
|
| 122 |
+
# X_features, info_df = get_descriptors(internal_df)
|
| 123 |
+
|
| 124 |
+
# Select descriptors
|
| 125 |
+
X_features = select_descriptors(df=X_features)
|
| 126 |
+
|
| 127 |
+
# Scale descriptors
|
| 128 |
+
X_features = scale_descriptors(df=X_features)
|
| 129 |
+
|
| 130 |
+
# Get classifier
|
| 131 |
+
# clf = get_clf(clf_str=clf, sampling_str=sampling)
|
| 132 |
+
|
| 133 |
+
# Get classifier
|
| 134 |
+
result_df = predict_permeability(clf_str=clf,
|
| 135 |
+
sampling_str=sampling,
|
| 136 |
+
features_df=X_features,
|
| 137 |
+
info_df=info_df,
|
| 138 |
+
threshold=threshold)
|
| 139 |
+
|
| 140 |
+
# Get classifier
|
| 141 |
+
display_cols = ["ID", "SMILES", "B3clf_predicted_probability", "B3clf_predicted_label"]
|
| 142 |
+
|
| 143 |
+
result_df = result_df[[col for col in result_df.columns.to_list() if col in display_cols]]
|
| 144 |
+
if verbose != 0:
|
| 145 |
+
print(result_df)
|
| 146 |
+
|
| 147 |
+
result_df.to_excel(output, index=None, engine="openpyxl")
|
| 148 |
+
|
| 149 |
+
if keep_features != "yes":
|
| 150 |
+
os.remove(features_out)
|
| 151 |
+
if keep_sdf != "yes":
|
| 152 |
+
os.remove(internal_sdf)
|
| 153 |
+
|
| 154 |
+
return result_df
|
b3clf/b3clf_structure.png
ADDED
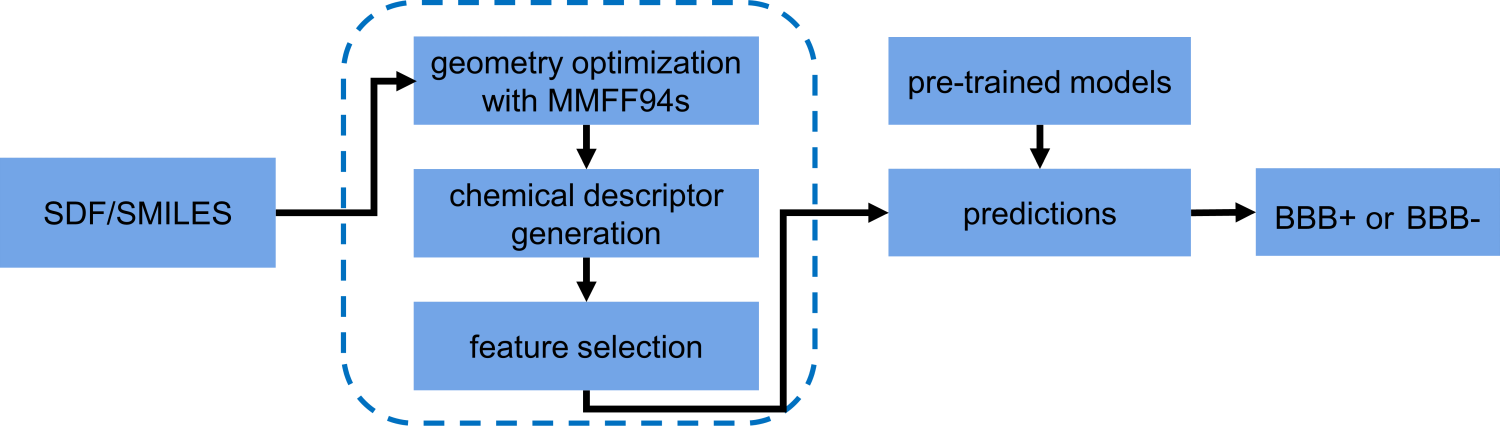
|
b3clf/data/B3clf_thresholds.xlsx
ADDED
|
Binary file (7.38 kB). View file
|
|
|
b3clf/descriptor_padel.py
ADDED
|
@@ -0,0 +1,90 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
import os
|
| 25 |
+
import sys
|
| 26 |
+
|
| 27 |
+
cwd = os.path.dirname(os.path.abspath(__file__))
|
| 28 |
+
sys.path.append(os.path.join(cwd, "padelpy"))
|
| 29 |
+
|
| 30 |
+
import pandas as pd
|
| 31 |
+
from rdkit import Chem
|
| 32 |
+
from padelpy import from_sdf
|
| 33 |
+
|
| 34 |
+
"""Compute PaDEL descriptors."""
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
def compute_descriptors(sdf_file,
|
| 38 |
+
# Change this to be an optional argument
|
| 39 |
+
excel_out="padel_descriptors.xlsx",
|
| 40 |
+
output_csv=None,
|
| 41 |
+
timeout=None,
|
| 42 |
+
time_per_molecule=-1,
|
| 43 |
+
) -> pd.DataFrame:
|
| 44 |
+
"""Compute the chemical descriptors with PaDEL.
|
| 45 |
+
|
| 46 |
+
Parameters
|
| 47 |
+
----------
|
| 48 |
+
sdf_file : str
|
| 49 |
+
Input SDF file name.
|
| 50 |
+
excel_out : str, optional
|
| 51 |
+
Excel file name to save PaDEL descriptors.
|
| 52 |
+
timeout : float
|
| 53 |
+
The maximum time, in seconds, for calculating the descriptors. When set to be None,
|
| 54 |
+
this does not take effect.
|
| 55 |
+
|
| 56 |
+
Returns
|
| 57 |
+
-------
|
| 58 |
+
df_desc : pandas.dataframe
|
| 59 |
+
The computed pandas dataframe of PaDEL descriptors.
|
| 60 |
+
|
| 61 |
+
"""
|
| 62 |
+
desc = from_sdf(sdf_file=sdf_file,
|
| 63 |
+
output_csv=output_csv,
|
| 64 |
+
descriptors=True,
|
| 65 |
+
fingerprints=False,
|
| 66 |
+
timeout=timeout,
|
| 67 |
+
maxruntime=time_per_molecule,
|
| 68 |
+
)
|
| 69 |
+
df_desc = pd.DataFrame(desc)
|
| 70 |
+
|
| 71 |
+
# add molecule names to dataframe
|
| 72 |
+
suppl = Chem.SDMolSupplier(sdf_file,
|
| 73 |
+
sanitize=True,
|
| 74 |
+
removeHs=False,
|
| 75 |
+
strictParsing=True)
|
| 76 |
+
mol_names = [mol.GetProp("_Name") for mol in suppl]
|
| 77 |
+
df_desc.index = mol_names
|
| 78 |
+
df_desc.index.name = "ID"
|
| 79 |
+
|
| 80 |
+
# drop rows with nan values
|
| 81 |
+
# todo: add imputation option
|
| 82 |
+
df_desc.dropna(axis=0, inplace=True)
|
| 83 |
+
|
| 84 |
+
# save results
|
| 85 |
+
if excel_out is not None:
|
| 86 |
+
df_desc.to_excel(excel_out, engine="openpyxl")
|
| 87 |
+
|
| 88 |
+
return df_desc
|
| 89 |
+
|
| 90 |
+
# Index will be the molecule's name
|
b3clf/feature_list.txt
ADDED
|
@@ -0,0 +1,475 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
nAcid
|
| 2 |
+
ALogP
|
| 3 |
+
ALogp2
|
| 4 |
+
AMR
|
| 5 |
+
naAromAtom
|
| 6 |
+
nH
|
| 7 |
+
nN
|
| 8 |
+
nO
|
| 9 |
+
nS
|
| 10 |
+
nP
|
| 11 |
+
nF
|
| 12 |
+
nCl
|
| 13 |
+
nBr
|
| 14 |
+
nI
|
| 15 |
+
nX
|
| 16 |
+
ATS0m
|
| 17 |
+
ATS4m
|
| 18 |
+
ATS2s
|
| 19 |
+
AATS0m
|
| 20 |
+
AATS1m
|
| 21 |
+
AATS4m
|
| 22 |
+
AATS5m
|
| 23 |
+
AATS6m
|
| 24 |
+
AATS7m
|
| 25 |
+
AATS8m
|
| 26 |
+
AATS0v
|
| 27 |
+
AATS3v
|
| 28 |
+
AATS5v
|
| 29 |
+
AATS7v
|
| 30 |
+
AATS0e
|
| 31 |
+
AATS4e
|
| 32 |
+
AATS5e
|
| 33 |
+
AATS6e
|
| 34 |
+
AATS7e
|
| 35 |
+
AATS4p
|
| 36 |
+
AATS0i
|
| 37 |
+
AATS1i
|
| 38 |
+
AATS4i
|
| 39 |
+
AATS5i
|
| 40 |
+
AATS8i
|
| 41 |
+
AATS4s
|
| 42 |
+
AATS5s
|
| 43 |
+
AATS6s
|
| 44 |
+
AATS7s
|
| 45 |
+
AATS8s
|
| 46 |
+
ATSC2c
|
| 47 |
+
ATSC3c
|
| 48 |
+
ATSC4c
|
| 49 |
+
ATSC5c
|
| 50 |
+
ATSC6c
|
| 51 |
+
ATSC7c
|
| 52 |
+
ATSC8c
|
| 53 |
+
ATSC1m
|
| 54 |
+
ATSC2m
|
| 55 |
+
ATSC3m
|
| 56 |
+
ATSC4m
|
| 57 |
+
ATSC5m
|
| 58 |
+
ATSC6m
|
| 59 |
+
ATSC7m
|
| 60 |
+
ATSC8m
|
| 61 |
+
ATSC1v
|
| 62 |
+
ATSC2v
|
| 63 |
+
ATSC3v
|
| 64 |
+
ATSC4v
|
| 65 |
+
ATSC5v
|
| 66 |
+
ATSC6v
|
| 67 |
+
ATSC7v
|
| 68 |
+
ATSC8v
|
| 69 |
+
ATSC1e
|
| 70 |
+
ATSC2e
|
| 71 |
+
ATSC3e
|
| 72 |
+
ATSC4e
|
| 73 |
+
ATSC5e
|
| 74 |
+
ATSC6e
|
| 75 |
+
ATSC7e
|
| 76 |
+
ATSC8e
|
| 77 |
+
ATSC1i
|
| 78 |
+
ATSC2i
|
| 79 |
+
ATSC3i
|
| 80 |
+
ATSC4i
|
| 81 |
+
ATSC5i
|
| 82 |
+
ATSC6i
|
| 83 |
+
ATSC7i
|
| 84 |
+
ATSC8i
|
| 85 |
+
ATSC1s
|
| 86 |
+
ATSC3s
|
| 87 |
+
AATSC1c
|
| 88 |
+
AATSC4c
|
| 89 |
+
AATSC5c
|
| 90 |
+
AATSC6c
|
| 91 |
+
AATSC7c
|
| 92 |
+
AATSC8c
|
| 93 |
+
AATSC2m
|
| 94 |
+
AATSC3m
|
| 95 |
+
AATSC4m
|
| 96 |
+
AATSC5m
|
| 97 |
+
AATSC6m
|
| 98 |
+
AATSC7m
|
| 99 |
+
AATSC8m
|
| 100 |
+
AATSC0v
|
| 101 |
+
AATSC4v
|
| 102 |
+
AATSC5v
|
| 103 |
+
AATSC6v
|
| 104 |
+
AATSC7v
|
| 105 |
+
AATSC8v
|
| 106 |
+
AATSC3e
|
| 107 |
+
AATSC4e
|
| 108 |
+
AATSC5e
|
| 109 |
+
AATSC6e
|
| 110 |
+
AATSC7e
|
| 111 |
+
AATSC8e
|
| 112 |
+
AATSC1p
|
| 113 |
+
AATSC2p
|
| 114 |
+
AATSC3p
|
| 115 |
+
AATSC4p
|
| 116 |
+
AATSC3i
|
| 117 |
+
AATSC4i
|
| 118 |
+
AATSC5i
|
| 119 |
+
AATSC6i
|
| 120 |
+
AATSC7i
|
| 121 |
+
AATSC8i
|
| 122 |
+
AATSC1s
|
| 123 |
+
AATSC2s
|
| 124 |
+
AATSC3s
|
| 125 |
+
AATSC4s
|
| 126 |
+
AATSC6s
|
| 127 |
+
AATSC8s
|
| 128 |
+
MATS1c
|
| 129 |
+
MATS3c
|
| 130 |
+
MATS3m
|
| 131 |
+
MATS4m
|
| 132 |
+
MATS5m
|
| 133 |
+
MATS6m
|
| 134 |
+
MATS1e
|
| 135 |
+
MATS3e
|
| 136 |
+
MATS1s
|
| 137 |
+
MATS2s
|
| 138 |
+
MATS3s
|
| 139 |
+
MATS4s
|
| 140 |
+
MATS5s
|
| 141 |
+
MATS7s
|
| 142 |
+
GATS1c
|
| 143 |
+
GATS2c
|
| 144 |
+
GATS3c
|
| 145 |
+
GATS4c
|
| 146 |
+
GATS5c
|
| 147 |
+
GATS6c
|
| 148 |
+
GATS7c
|
| 149 |
+
GATS8c
|
| 150 |
+
GATS1m
|
| 151 |
+
GATS2m
|
| 152 |
+
GATS4m
|
| 153 |
+
GATS5m
|
| 154 |
+
GATS6m
|
| 155 |
+
GATS7m
|
| 156 |
+
GATS8m
|
| 157 |
+
GATS2v
|
| 158 |
+
GATS6v
|
| 159 |
+
GATS7v
|
| 160 |
+
GATS8v
|
| 161 |
+
GATS1e
|
| 162 |
+
GATS3e
|
| 163 |
+
GATS4e
|
| 164 |
+
GATS1p
|
| 165 |
+
GATS3p
|
| 166 |
+
GATS4p
|
| 167 |
+
GATS5p
|
| 168 |
+
GATS1i
|
| 169 |
+
GATS2i
|
| 170 |
+
GATS3i
|
| 171 |
+
GATS4i
|
| 172 |
+
GATS2s
|
| 173 |
+
GATS3s
|
| 174 |
+
GATS5s
|
| 175 |
+
GATS6s
|
| 176 |
+
GATS7s
|
| 177 |
+
GATS8s
|
| 178 |
+
SM1_DzZ
|
| 179 |
+
VE1_DzZ
|
| 180 |
+
VE3_DzZ
|
| 181 |
+
VR1_DzZ
|
| 182 |
+
VE1_Dzv
|
| 183 |
+
VR2_Dzv
|
| 184 |
+
VE2_Dze
|
| 185 |
+
SpMAD_Dzp
|
| 186 |
+
VE2_Dzp
|
| 187 |
+
SpMAD_Dzs
|
| 188 |
+
SM1_Dzs
|
| 189 |
+
VE1_Dzs
|
| 190 |
+
VR2_Dzs
|
| 191 |
+
nBase
|
| 192 |
+
BCUTw-1l
|
| 193 |
+
BCUTw-1h
|
| 194 |
+
BCUTc-1l
|
| 195 |
+
BCUTc-1h
|
| 196 |
+
BCUTp-1l
|
| 197 |
+
BCUTp-1h
|
| 198 |
+
nBondsD
|
| 199 |
+
nBondsD2
|
| 200 |
+
nBondsT
|
| 201 |
+
SpMax3_Bhm
|
| 202 |
+
SpMin1_Bhm
|
| 203 |
+
SpMin2_Bhm
|
| 204 |
+
SpMin4_Bhm
|
| 205 |
+
SpMin7_Bhm
|
| 206 |
+
SpMax1_Bhv
|
| 207 |
+
SpMax2_Bhe
|
| 208 |
+
SpMin8_Bhe
|
| 209 |
+
SpMax1_Bhs
|
| 210 |
+
SpMax3_Bhs
|
| 211 |
+
C2SP1
|
| 212 |
+
C1SP2
|
| 213 |
+
C2SP2
|
| 214 |
+
C3SP2
|
| 215 |
+
C1SP3
|
| 216 |
+
C2SP3
|
| 217 |
+
C3SP3
|
| 218 |
+
C4SP3
|
| 219 |
+
SCH-3
|
| 220 |
+
SCH-4
|
| 221 |
+
SCH-5
|
| 222 |
+
SCH-7
|
| 223 |
+
SC-4
|
| 224 |
+
SC-5
|
| 225 |
+
VC-3
|
| 226 |
+
SP-7
|
| 227 |
+
ASP-0
|
| 228 |
+
ASP-1
|
| 229 |
+
ASP-4
|
| 230 |
+
ASP-5
|
| 231 |
+
ASP-6
|
| 232 |
+
ASP-7
|
| 233 |
+
AVP-0
|
| 234 |
+
AVP-3
|
| 235 |
+
AVP-4
|
| 236 |
+
AVP-6
|
| 237 |
+
AVP-7
|
| 238 |
+
CrippenLogP
|
| 239 |
+
VE1_Dt
|
| 240 |
+
VE3_Dt
|
| 241 |
+
VR1_Dt
|
| 242 |
+
ECCEN
|
| 243 |
+
nwHBd
|
| 244 |
+
nHBint2
|
| 245 |
+
nHBint3
|
| 246 |
+
nHBint4
|
| 247 |
+
nHBint5
|
| 248 |
+
nHBint6
|
| 249 |
+
nHBint7
|
| 250 |
+
nHBint8
|
| 251 |
+
nHBint9
|
| 252 |
+
nHdNH
|
| 253 |
+
nHsSH
|
| 254 |
+
nHsNH2
|
| 255 |
+
nHssNH
|
| 256 |
+
nHaaNH
|
| 257 |
+
nHtCH
|
| 258 |
+
nHdCH2
|
| 259 |
+
nHdsCH
|
| 260 |
+
nHCsatu
|
| 261 |
+
nHAvin
|
| 262 |
+
nsCH3
|
| 263 |
+
nssCH2
|
| 264 |
+
nsssCH
|
| 265 |
+
naaaC
|
| 266 |
+
ndsN
|
| 267 |
+
naaN
|
| 268 |
+
nsssN
|
| 269 |
+
naasN
|
| 270 |
+
nssssNp
|
| 271 |
+
ndO
|
| 272 |
+
nssO
|
| 273 |
+
naaO
|
| 274 |
+
nsOm
|
| 275 |
+
ndS
|
| 276 |
+
naaS
|
| 277 |
+
ndssS
|
| 278 |
+
nddssS
|
| 279 |
+
SwHBa
|
| 280 |
+
SHBint10
|
| 281 |
+
SHsOH
|
| 282 |
+
SsssCH
|
| 283 |
+
SdssC
|
| 284 |
+
SaasC
|
| 285 |
+
SssssC
|
| 286 |
+
SssS
|
| 287 |
+
SsBr
|
| 288 |
+
SsI
|
| 289 |
+
minHBd
|
| 290 |
+
minHBa
|
| 291 |
+
minwHBa
|
| 292 |
+
minHBint2
|
| 293 |
+
minHBint3
|
| 294 |
+
minHBint4
|
| 295 |
+
minHBint5
|
| 296 |
+
minHBint6
|
| 297 |
+
minHBint7
|
| 298 |
+
minHBint8
|
| 299 |
+
minHBint9
|
| 300 |
+
minHBint10
|
| 301 |
+
minHsOH
|
| 302 |
+
minHsNH2
|
| 303 |
+
minHssNH
|
| 304 |
+
minHdsCH
|
| 305 |
+
minHaaCH
|
| 306 |
+
minHCsats
|
| 307 |
+
minHCsatu
|
| 308 |
+
minHother
|
| 309 |
+
minsCH3
|
| 310 |
+
minssCH2
|
| 311 |
+
minsssCH
|
| 312 |
+
minaasC
|
| 313 |
+
mintN
|
| 314 |
+
minsssN
|
| 315 |
+
minsOH
|
| 316 |
+
mindO
|
| 317 |
+
minssO
|
| 318 |
+
minsF
|
| 319 |
+
maxHBa
|
| 320 |
+
maxwHBa
|
| 321 |
+
maxHBint2
|
| 322 |
+
maxHBint3
|
| 323 |
+
maxHBint4
|
| 324 |
+
maxHBint5
|
| 325 |
+
maxHBint6
|
| 326 |
+
maxHBint7
|
| 327 |
+
maxHBint9
|
| 328 |
+
maxHBint10
|
| 329 |
+
maxHCsats
|
| 330 |
+
maxssCH2
|
| 331 |
+
maxsssCH
|
| 332 |
+
maxdssC
|
| 333 |
+
maxssssC
|
| 334 |
+
maxsI
|
| 335 |
+
hmax
|
| 336 |
+
hmin
|
| 337 |
+
ETA_AlphaP
|
| 338 |
+
ETA_dAlpha_A
|
| 339 |
+
ETA_dEpsilon_B
|
| 340 |
+
ETA_dEpsilon_D
|
| 341 |
+
ETA_dPsi_B
|
| 342 |
+
ETA_Shape_Y
|
| 343 |
+
ETA_BetaP_s
|
| 344 |
+
ETA_Beta_ns_d
|
| 345 |
+
ETA_EtaP_B
|
| 346 |
+
IC0
|
| 347 |
+
IC1
|
| 348 |
+
IC2
|
| 349 |
+
SIC1
|
| 350 |
+
SIC3
|
| 351 |
+
SIC5
|
| 352 |
+
BIC0
|
| 353 |
+
MIC5
|
| 354 |
+
ZMIC2
|
| 355 |
+
ZMIC5
|
| 356 |
+
Kier3
|
| 357 |
+
nAtomLC
|
| 358 |
+
nAtomP
|
| 359 |
+
nAtomLAC
|
| 360 |
+
MDEC-14
|
| 361 |
+
MDEC-22
|
| 362 |
+
MDEC-23
|
| 363 |
+
MDEC-33
|
| 364 |
+
MDEO-11
|
| 365 |
+
MDEO-12
|
| 366 |
+
MDEN-11
|
| 367 |
+
MDEN-12
|
| 368 |
+
MDEN-13
|
| 369 |
+
MDEN-22
|
| 370 |
+
MDEN-23
|
| 371 |
+
MDEN-33
|
| 372 |
+
MLFER_A
|
| 373 |
+
MLFER_BH
|
| 374 |
+
MLFER_S
|
| 375 |
+
piPC10
|
| 376 |
+
R_TpiPCTPC
|
| 377 |
+
PetitjeanNumber
|
| 378 |
+
n5Ring
|
| 379 |
+
n6Ring
|
| 380 |
+
n7Ring
|
| 381 |
+
n8Ring
|
| 382 |
+
n12Ring
|
| 383 |
+
nG12Ring
|
| 384 |
+
nFRing
|
| 385 |
+
nF4Ring
|
| 386 |
+
nF6Ring
|
| 387 |
+
nF7Ring
|
| 388 |
+
nF8Ring
|
| 389 |
+
nF9Ring
|
| 390 |
+
nF10Ring
|
| 391 |
+
nF11Ring
|
| 392 |
+
nF12Ring
|
| 393 |
+
nT7Ring
|
| 394 |
+
nHeteroRing
|
| 395 |
+
n3HeteroRing
|
| 396 |
+
n6HeteroRing
|
| 397 |
+
n8HeteroRing
|
| 398 |
+
nF6HeteroRing
|
| 399 |
+
nF10HeteroRing
|
| 400 |
+
RotBFrac
|
| 401 |
+
nRotBt
|
| 402 |
+
LipinskiFailures
|
| 403 |
+
topoRadius
|
| 404 |
+
JGI2
|
| 405 |
+
JGI3
|
| 406 |
+
JGI4
|
| 407 |
+
JGI5
|
| 408 |
+
JGI6
|
| 409 |
+
JGI7
|
| 410 |
+
JGI8
|
| 411 |
+
JGI9
|
| 412 |
+
JGI10
|
| 413 |
+
VE1_D
|
| 414 |
+
VE3_D
|
| 415 |
+
VR1_D
|
| 416 |
+
SRW9
|
| 417 |
+
TDB1u
|
| 418 |
+
TDB4u
|
| 419 |
+
TDB5u
|
| 420 |
+
TDB9u
|
| 421 |
+
TDB10u
|
| 422 |
+
TDB9m
|
| 423 |
+
TDB10m
|
| 424 |
+
TDB9v
|
| 425 |
+
TDB2i
|
| 426 |
+
TDB9s
|
| 427 |
+
TDB10s
|
| 428 |
+
PPSA-3
|
| 429 |
+
DPSA-1
|
| 430 |
+
FPSA-3
|
| 431 |
+
FNSA-3
|
| 432 |
+
RPCG
|
| 433 |
+
RNCG
|
| 434 |
+
RPCS
|
| 435 |
+
RNCS
|
| 436 |
+
THSA
|
| 437 |
+
LOBMAX
|
| 438 |
+
MOMI-Y
|
| 439 |
+
MOMI-XY
|
| 440 |
+
geomShape
|
| 441 |
+
RDF20u
|
| 442 |
+
RDF100u
|
| 443 |
+
RDF155u
|
| 444 |
+
RDF10m
|
| 445 |
+
RDF20m
|
| 446 |
+
RDF35m
|
| 447 |
+
RDF40m
|
| 448 |
+
RDF55m
|
| 449 |
+
RDF60m
|
| 450 |
+
RDF65m
|
| 451 |
+
RDF110m
|
| 452 |
+
RDF125m
|
| 453 |
+
RDF130m
|
| 454 |
+
RDF135m
|
| 455 |
+
RDF140m
|
| 456 |
+
RDF30p
|
| 457 |
+
RDF40s
|
| 458 |
+
RDF80s
|
| 459 |
+
RDF115s
|
| 460 |
+
RDF145s
|
| 461 |
+
L2u
|
| 462 |
+
L3u
|
| 463 |
+
P1u
|
| 464 |
+
E1u
|
| 465 |
+
E2u
|
| 466 |
+
E3u
|
| 467 |
+
Du
|
| 468 |
+
E1m
|
| 469 |
+
E2m
|
| 470 |
+
E3m
|
| 471 |
+
Dm
|
| 472 |
+
E1v
|
| 473 |
+
E2v
|
| 474 |
+
E3v
|
| 475 |
+
Dv
|
b3clf/geometry_opt.py
ADDED
|
@@ -0,0 +1,208 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
import pandas as pd
|
| 25 |
+
from rdkit import Chem
|
| 26 |
+
from rdkit.Chem import AllChem
|
| 27 |
+
|
| 28 |
+
"""Convert SMILES to 3D and/or minimize the geometry from SDF with force field."""
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
def geometry_optimize(input_fname,
|
| 32 |
+
output_sdf,
|
| 33 |
+
steps_opt=10000,
|
| 34 |
+
# convergence=1.e-7,
|
| 35 |
+
tool="rdkit",
|
| 36 |
+
# optimization="cg",
|
| 37 |
+
force_field="MMFF94s",
|
| 38 |
+
smi_col=None,
|
| 39 |
+
sep="\s+|t+"):
|
| 40 |
+
"""Generate 3D coordinates and run geometry optimization with force field."""
|
| 41 |
+
|
| 42 |
+
# optimize the 3d coordinates
|
| 43 |
+
# use RDKit to minimize the geometry
|
| 44 |
+
if tool.lower() == "rdkit":
|
| 45 |
+
minimize_with_rdkit(input_molfname=input_fname,
|
| 46 |
+
sdf_out=output_sdf,
|
| 47 |
+
maxIters=steps_opt,
|
| 48 |
+
force_field=force_field,
|
| 49 |
+
smi_col=smi_col,
|
| 50 |
+
sep=sep)
|
| 51 |
+
# use openbabel to minimize the geometry
|
| 52 |
+
elif tool == "openbabel":
|
| 53 |
+
# minimize_with_openbabel(input_molfname=input_fname,
|
| 54 |
+
# sdf_out=output_sdf,
|
| 55 |
+
# steps=steps_opt,
|
| 56 |
+
# optimization=optimization,
|
| 57 |
+
# convergence=convergence,
|
| 58 |
+
# force_field=force_field,
|
| 59 |
+
# smi_col=smi_col)
|
| 60 |
+
raise ValueError("OpenBabel is not supported yet.")
|
| 61 |
+
else:
|
| 62 |
+
raise ValueError("{} not implemented yet.".format(tool))
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
def minimize_with_rdkit(input_molfname,
|
| 66 |
+
sdf_out,
|
| 67 |
+
smi_col=None,
|
| 68 |
+
mol_name_col=None,
|
| 69 |
+
maxIters=400,
|
| 70 |
+
force_field="MMFF94s",
|
| 71 |
+
sep="\s+"):
|
| 72 |
+
"""Add hydrogen for 3D coordinates and minimize the geometry with RdKit."""
|
| 73 |
+
# load molecules
|
| 74 |
+
if input_molfname.lower().endswith(".smi") or input_molfname.lower().endswith(".csv"):
|
| 75 |
+
# todo: support .txt files
|
| 76 |
+
# todo: add support of more flexible separators
|
| 77 |
+
# todo: fix problem when mol_name is empty
|
| 78 |
+
df_mol = pd.read_csv(input_molfname, sep=sep, engine="python", header=None)
|
| 79 |
+
if df_mol.shape[1] == 1:
|
| 80 |
+
# Case for only SMILES column
|
| 81 |
+
smile_list = df_mol.iloc[:, -1].to_list()
|
| 82 |
+
mol_name_list = df_mol.iloc[:, -1].to_list()
|
| 83 |
+
else:
|
| 84 |
+
# Case for SMILES and MOL name columns
|
| 85 |
+
if smi_col is None:
|
| 86 |
+
smile_list = df_mol.iloc[:, 0].to_list()
|
| 87 |
+
else:
|
| 88 |
+
smile_list = df_mol[smi_col].to_list()
|
| 89 |
+
|
| 90 |
+
if mol_name_col is None:
|
| 91 |
+
# todo: use name if column name is valid
|
| 92 |
+
mol_name_list = df_mol.iloc[:, -1].to_list()
|
| 93 |
+
else:
|
| 94 |
+
mol_name_list = df_mol[mol_name_col].to_list()
|
| 95 |
+
|
| 96 |
+
mols = []
|
| 97 |
+
for idx, smi in enumerate(smile_list):
|
| 98 |
+
mol = Chem.MolFromSmiles(smi)
|
| 99 |
+
# This will overwrite
|
| 100 |
+
if mol is not None:
|
| 101 |
+
mol.SetProp("_Name", mol_name_list[idx])
|
| 102 |
+
mols.append(mol)
|
| 103 |
+
|
| 104 |
+
elif input_molfname.lower().endswith(".sdf"):
|
| 105 |
+
suppl = Chem.SDMolSupplier(input_molfname,
|
| 106 |
+
sanitize=True,
|
| 107 |
+
removeHs=False,
|
| 108 |
+
strictParsing=True)
|
| 109 |
+
mols = [mol for mol in suppl]
|
| 110 |
+
for idx, mol in enumerate(mols):
|
| 111 |
+
if (mol.GetProp("_Name") == "") or (mol.GetProp("_Name") is None):
|
| 112 |
+
smi = Chem.MolToSmiles(mol)
|
| 113 |
+
mol.SetProp("_Name", smi)
|
| 114 |
+
mols[idx] = mol
|
| 115 |
+
|
| 116 |
+
writer = Chem.SDWriter(sdf_out)
|
| 117 |
+
for idx, mol in enumerate(mols):
|
| 118 |
+
mol = Chem.AddHs(mol)
|
| 119 |
+
if force_field == "MMFF94s":
|
| 120 |
+
# use MMFF~ force field if possible
|
| 121 |
+
|
| 122 |
+
# taken from
|
| 123 |
+
# https://open-babel.readthedocs.io/en/latest/Forcefields/mmff94.html
|
| 124 |
+
# Some experiments and most theoretical calculations show significant pyramidal
|
| 125 |
+
# “puckering” at nitrogens in isolated structures. The MMFF94s (static) variant has
|
| 126 |
+
# slightly different out-of-plane bending and dihedral torsion parameters to planarize
|
| 127 |
+
# certain types of delocalized trigonal N atoms, such as aromatic aniline. This provides
|
| 128 |
+
# a better match to the time-average molecular geometry in solution or crystal
|
| 129 |
+
# structures.
|
| 130 |
+
#
|
| 131 |
+
# If you are comparing force-field optimized molecules to crystal structure geometries,
|
| 132 |
+
# we recommend using the MMFF94s variant for this reason. All other parameters are
|
| 133 |
+
# identical. However, if you are performing “docking” simulations, consideration of
|
| 134 |
+
# active solution conformations, or other types of computational studies, we recommend
|
| 135 |
+
# using the MMFF94 variant, since one form or another of the N geometry will
|
| 136 |
+
# predominate.
|
| 137 |
+
|
| 138 |
+
AllChem.EmbedMolecule(mol, randomSeed=999)
|
| 139 |
+
# the following code will raise some errors
|
| 140 |
+
mini_tag = AllChem.MMFFOptimizeMolecule(mol, force_field, maxIters=maxIters)
|
| 141 |
+
# 0 optimize converged
|
| 142 |
+
# -1 can not set up force field
|
| 143 |
+
# 1 more iterations required
|
| 144 |
+
if mini_tag == 0:
|
| 145 |
+
writer.write(mol)
|
| 146 |
+
else:
|
| 147 |
+
if mini_tag == 1:
|
| 148 |
+
AllChem.MMFFOptimizeMolecule(mol, force_field, maxIters=maxIters * 2)
|
| 149 |
+
elif mini_tag == -1:
|
| 150 |
+
AllChem.UFFOptimizeMolecule(mol, maxIters=400)
|
| 151 |
+
writer.write(mol)
|
| 152 |
+
|
| 153 |
+
elif force_field == "uff":
|
| 154 |
+
# use uff force field if possible
|
| 155 |
+
AllChem.EmbedMolecule(mol, randomSeed=999)
|
| 156 |
+
# the following code will raise some errors
|
| 157 |
+
mini_tag = AllChem.UFFOptimizeMolecule(mol, maxIters=maxIters)
|
| 158 |
+
# 0 optimize converged
|
| 159 |
+
# -1 can not set up force field
|
| 160 |
+
# 1 more iterations required
|
| 161 |
+
if mini_tag == 0:
|
| 162 |
+
writer.write(mol)
|
| 163 |
+
else:
|
| 164 |
+
if mini_tag == 1:
|
| 165 |
+
AllChem.UFFOptimizeMolecule(mol, maxIters=maxIters * 2)
|
| 166 |
+
elif mini_tag == -1:
|
| 167 |
+
AllChem.MMFFOptimizeMolecule(mol, "MMFF94s", maxIters=maxIters)
|
| 168 |
+
writer.write(mol)
|
| 169 |
+
|
| 170 |
+
else:
|
| 171 |
+
raise NotImplementedError("This method is not implemented yet.")
|
| 172 |
+
|
| 173 |
+
writer.close()
|
| 174 |
+
|
| 175 |
+
# todo: now the implementation is not supporting adding molecule name (such as SMILES strings)
|
| 176 |
+
# def minimize_with_openbabel(input_molfname,
|
| 177 |
+
# sdf_out,
|
| 178 |
+
# steps=10000,
|
| 179 |
+
# convergence=1.e-7,
|
| 180 |
+
# optimization="cg",
|
| 181 |
+
# force_field="GAFF",
|
| 182 |
+
# smi_col=None):
|
| 183 |
+
# """Minimize the geometries with openbabel.
|
| 184 |
+
#
|
| 185 |
+
# Parameters
|
| 186 |
+
# ----------
|
| 187 |
+
# input_molfname : str
|
| 188 |
+
# Input molecule fie name.
|
| 189 |
+
# sdf_out : str
|
| 190 |
+
# Output molecule file name.
|
| 191 |
+
# steps : int, optional
|
| 192 |
+
# Specify the maximum number of steps. default=2500.
|
| 193 |
+
# optimization : str, optional
|
| 194 |
+
# Use conjugate gradients ("cg") or steepest descent ("sd") algorithm for optimization.
|
| 195 |
+
# Default="cg".
|
| 196 |
+
# convergence : float, optional
|
| 197 |
+
# convergence threshold. Default=1.e-7.
|
| 198 |
+
# force_field : str, optional
|
| 199 |
+
# ForceField name including Generalized Amber Force Field (gaff), Ghemical Force Field
|
| 200 |
+
# (ghemical), MMFF94 Force Field (mmff94) and Universal Force Field (uff). Default="gaff".
|
| 201 |
+
# """
|
| 202 |
+
#
|
| 203 |
+
# # https://open-babel.readthedocs.io/en/latest/Command-line_tools/babel.html#forcefield-energy-and-minimization
|
| 204 |
+
# subprocess.Popen(["obabel", input_molfname, "-h", "-O", sdf_out,
|
| 205 |
+
# "--gen3d", "--minimize",
|
| 206 |
+
# "--n", str(steps), "--sd", optimization, "--crit",
|
| 207 |
+
# str(convergence), "--ff", force_field])
|
| 208 |
+
# print("Geometry optimization with OpenBabel is done.")
|
b3clf/pre_trained/b3clf_dtree_borderline_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:16acf21e0ddf7185a35a101c5c981c3bc55b8b00f4178051ff30e98078e5f7cd
|
| 3 |
+
size 26214
|
b3clf/pre_trained/b3clf_dtree_classic_ADASYN.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0bfa4680b3c52dafecf081e3cf957265ba787994bc83d3babb157157f7887388
|
| 3 |
+
size 21033
|
b3clf/pre_trained/b3clf_dtree_classic_RandUndersampling.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6abf2ba341914920dccc5632825374187855a32e3b8f96fcc01d1cdc789a776f
|
| 3 |
+
size 15269
|
b3clf/pre_trained/b3clf_dtree_classic_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:3a13c59aea7c4f6bd14f4c8bf2baf1b46b933cfd5992e282c54e989dffa51486
|
| 3 |
+
size 18293
|
b3clf/pre_trained/b3clf_dtree_common.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6f411314a26165f42ac654a3744a3be82b3437e40b4a88b8e1f04250800707c2
|
| 3 |
+
size 13254
|
b3clf/pre_trained/b3clf_dtree_kmeans_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:9f6cce007cd59a7589dbc1408771b5d71cc1f931bf6cea98e533d9e8617f76a8
|
| 3 |
+
size 28379
|
b3clf/pre_trained/b3clf_knn_borderline_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:19c1a0da174893b44d3127198311672f87e766b74584e7108264a5b321ffe73b
|
| 3 |
+
size 36976533
|
b3clf/pre_trained/b3clf_knn_classic_ADASYN.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:470995041fbe430a33e7eebf9fa9cc3c8ac3876e157773748397ac8caea03b9e
|
| 3 |
+
size 75157179
|
b3clf/pre_trained/b3clf_knn_classic_RandUndersampling.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c62c4bc3d20e6e627be7fd7b1e1e2aa758ff082030c16a33d9aabb9c01925e11
|
| 3 |
+
size 38992348
|
b3clf/pre_trained/b3clf_knn_classic_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f27fbcfd884a1821ab580d1a94d0f84f5158ccacb3cd7ce17fe93a17f6fa82d7
|
| 3 |
+
size 36976533
|
b3clf/pre_trained/b3clf_knn_common.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0da2fa65aca1bc7fb1a0da229692af238b1c7d22e5117e7f36dc6837a1c9e8c1
|
| 3 |
+
size 56899868
|
b3clf/pre_trained/b3clf_knn_kmeans_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:027c788f29d994ea3086538550c969f22b13f0f61521f302870611182d4f4be3
|
| 3 |
+
size 36991774
|
b3clf/pre_trained/b3clf_logreg_borderline_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:ce4cc05e90dd86d98e2ea92aa147d2aa26b9ac09918f86c7d1cd46a1fd9df925
|
| 3 |
+
size 4702
|
b3clf/pre_trained/b3clf_logreg_classic_ADASYN.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f84f5d05f266a5692d57f3aa4e0f3419e5a909588c70b16f8dbb39bafb75f98d
|
| 3 |
+
size 4702
|
b3clf/pre_trained/b3clf_logreg_classic_RandUndersampling.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a4d2b43613ef968b5b033c43c8d1e2af0ed1c93cebdda5095a23726dfcda3f74
|
| 3 |
+
size 4707
|
b3clf/pre_trained/b3clf_logreg_classic_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:3d09958c5037bd5aef31f0c84b8d6a48be14188dd16fb944ac49601a189ff162
|
| 3 |
+
size 4703
|
b3clf/pre_trained/b3clf_logreg_common.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f231476730bf5936a6b1c1e42594fcff4c5208629329d45dff83dca30bf3090a
|
| 3 |
+
size 4701
|
b3clf/pre_trained/b3clf_logreg_kmeans_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:17e75330a97d0aec1d079199b902afbd851881f0f85ac0d13e57a5f48896dee8
|
| 3 |
+
size 4702
|
b3clf/pre_trained/b3clf_scaler.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:382b16ac9ce81f10790f34d47aa10c5eab34f21025cc6660af4b3bb23b0ade3c
|
| 3 |
+
size 11998
|
b3clf/pre_trained/b3clf_xgb_borderline_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:2029f1933f6d31610c5be6eb9158e685f518458c501570ffc2802c7a8a508391
|
| 3 |
+
size 13918498
|
b3clf/pre_trained/b3clf_xgb_classic_ADASYN.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:43c39da319b24259a14a74dc4e77fc90727f10fcdc74d568191d6e444c95ccef
|
| 3 |
+
size 15434996
|
b3clf/pre_trained/b3clf_xgb_classic_RandUndersampling.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:eb4633a0ab2efbebd8f0b8e8f91e0063cdb751e6dbbfe5a6ebae8a14ea1b6aa1
|
| 3 |
+
size 8790808
|
b3clf/pre_trained/b3clf_xgb_classic_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:ca1e553e9105e6bae0d0de4643d9a110f279239c29f8304db891f9d2ad2ab1b9
|
| 3 |
+
size 13842083
|
b3clf/pre_trained/b3clf_xgb_common.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0d486e1c438b537b9c0a26329a634cb7c367863c96250daeee0b824a4294316b
|
| 3 |
+
size 8319534
|
b3clf/pre_trained/b3clf_xgb_kmeans_SMOTE.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8d5292c309a6ab6c8f4eefb006980f5b2484f5a14ca3280410dd2574287197a0
|
| 3 |
+
size 11061609
|
b3clf/test/test_SMILES.csv
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[H]OC(=O)C([H])([H])C([H])([H])C([H])([H])N1C([H])([H])C([H])([H])C([H])(OC([H])(c2nc([H])c([H])c([H])c2[H])c2c([H])c([H])c(Cl)c([H])c2[H])C([H])([H])C1([H])[H]
|
| 2 |
+
[H]OC(c1c([H])c([H])c([H])c([H])c1[H])(c1c([H])c([H])c([H])c([H])c1[H])C1([H])C([H])([H])N2C([H])([H])C([H])([H])C1([H])C([H])([H])C2([H])[H]
|
| 3 |
+
[H]c1nc2c(c([H])c1[H])C([H])([H])C([H])([H])c1c([H])c(Cl)c([H])c([H])c1C2=C1C([H])([H])C([H])([H])N(C([H])([H])c2c([H])nc([H])c(C([H])([H])[H])c2[H])C([H])([H])C1([H])[H]
|
| 4 |
+
CCC
|
| 5 |
+
CCCC
|
| 6 |
+
CC(=O)OC1=CC=CC=C1C(=O)O
|
| 7 |
+
CC(=O)OC1=C(C=C(C=C1)Cl)C(=O)OC(=O)C2=C(C=CC(=C2)Cl)OC(=O)C
|
b3clf/test/test_input_sdf.sdf
ADDED
|
@@ -0,0 +1,387 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
H1_Bepotastine
|
| 2 |
+
RDKit 3D
|
| 3 |
+
|
| 4 |
+
52 54 0 0 1 0 0 0 0 0999 V2000
|
| 5 |
+
6.2601 3.8627 -0.7580 Cl 0 0 0 0 0 0 0 0 0 0 0 0
|
| 6 |
+
0.7350 0.2169 -0.1032 O 0 0 0 0 0 0 0 0 0 0 0 0
|
| 7 |
+
-7.2627 2.0029 -1.7812 O 0 0 0 0 0 0 0 0 0 0 0 0
|
| 8 |
+
-7.8739 -0.0429 -1.1421 O 0 0 0 0 0 0 0 0 0 0 0 0
|
| 9 |
+
-3.2826 0.1387 1.0997 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 10 |
+
2.0420 -2.0119 -1.2138 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 11 |
+
-0.4341 -0.2713 0.5552 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 12 |
+
-1.5088 -0.5144 -0.4974 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 13 |
+
-0.9255 0.7694 1.5572 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 14 |
+
-2.8345 -0.8975 0.1550 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 15 |
+
-2.2740 0.3674 2.1479 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 16 |
+
-4.5811 -0.1850 1.7144 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 17 |
+
-5.7574 -0.2607 0.7330 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 18 |
+
1.9672 -0.2099 0.5040 C 0 0 2 0 0 0 0 0 0 0 0 0
|
| 19 |
+
-5.9298 1.0111 -0.0974 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 20 |
+
3.0410 0.8232 0.1855 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 21 |
+
2.3687 -1.6155 0.0463 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 22 |
+
3.9935 1.1819 1.1545 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 23 |
+
3.1185 1.4155 -1.0867 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 24 |
+
-7.1061 0.8976 -1.0266 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 25 |
+
3.0746 -2.4482 0.9176 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 26 |
+
4.9873 2.1194 0.8610 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 27 |
+
4.1084 2.3564 -1.3784 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 28 |
+
3.4496 -3.7187 0.4871 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 29 |
+
5.0380 2.7045 -0.4026 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 30 |
+
2.4252 -3.2455 -1.6060 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 31 |
+
3.1214 -4.1271 -0.7990 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 32 |
+
-0.2263 -1.2199 1.0679 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 33 |
+
-1.6364 0.3807 -1.1209 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 34 |
+
-1.1831 -1.3082 -1.1808 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 35 |
+
-0.1894 0.8975 2.3595 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 36 |
+
-1.0042 1.7496 1.0680 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 37 |
+
-3.5642 -1.0250 -0.6514 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 38 |
+
-2.7343 -1.8665 0.6611 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 39 |
+
-2.1498 -0.5299 2.7684 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 40 |
+
-2.6054 1.1766 2.8103 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 41 |
+
-4.5185 -1.1314 2.2673 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 42 |
+
-4.8272 0.5917 2.4507 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 43 |
+
-5.6514 -1.1306 0.0739 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 44 |
+
-6.6737 -0.4399 1.3108 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 45 |
+
1.8204 -0.2159 1.5927 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 46 |
+
-6.0945 1.8686 0.5639 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 47 |
+
-5.0396 1.1941 -0.7083 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 48 |
+
3.9687 0.7355 2.1458 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 49 |
+
2.3964 1.1402 -1.8552 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 50 |
+
3.3355 -2.1177 1.9176 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 51 |
+
5.7167 2.3889 1.6199 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 52 |
+
4.1451 2.8085 -2.3655 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 53 |
+
3.9993 -4.3824 1.1485 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 54 |
+
2.1492 -3.5132 -2.6219 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 55 |
+
3.4047 -5.1069 -1.1664 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 56 |
+
-8.0410 1.8004 -2.3409 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 57 |
+
1 25 1 0
|
| 58 |
+
2 7 1 0
|
| 59 |
+
2 14 1 0
|
| 60 |
+
3 20 1 0
|
| 61 |
+
3 52 1 0
|
| 62 |
+
4 20 2 0
|
| 63 |
+
5 10 1 0
|
| 64 |
+
5 11 1 0
|
| 65 |
+
5 12 1 0
|
| 66 |
+
6 17 2 0
|
| 67 |
+
6 26 1 0
|
| 68 |
+
7 8 1 0
|
| 69 |
+
7 9 1 0
|
| 70 |
+
7 28 1 0
|
| 71 |
+
8 10 1 0
|
| 72 |
+
8 29 1 0
|
| 73 |
+
8 30 1 0
|
| 74 |
+
9 11 1 0
|
| 75 |
+
9 31 1 0
|
| 76 |
+
9 32 1 0
|
| 77 |
+
10 33 1 0
|
| 78 |
+
10 34 1 0
|
| 79 |
+
11 35 1 0
|
| 80 |
+
11 36 1 0
|
| 81 |
+
12 13 1 0
|
| 82 |
+
12 37 1 0
|
| 83 |
+
12 38 1 0
|
| 84 |
+
13 15 1 0
|
| 85 |
+
13 39 1 0
|
| 86 |
+
13 40 1 0
|
| 87 |
+
14 16 1 0
|
| 88 |
+
14 17 1 0
|
| 89 |
+
14 41 1 1
|
| 90 |
+
15 20 1 0
|
| 91 |
+
15 42 1 0
|
| 92 |
+
15 43 1 0
|
| 93 |
+
16 18 2 0
|
| 94 |
+
16 19 1 0
|
| 95 |
+
17 21 1 0
|
| 96 |
+
18 22 1 0
|
| 97 |
+
18 44 1 0
|
| 98 |
+
19 23 2 0
|
| 99 |
+
19 45 1 0
|
| 100 |
+
21 24 2 0
|
| 101 |
+
21 46 1 0
|
| 102 |
+
22 25 2 0
|
| 103 |
+
22 47 1 0
|
| 104 |
+
23 25 1 0
|
| 105 |
+
23 48 1 0
|
| 106 |
+
24 27 1 0
|
| 107 |
+
24 49 1 0
|
| 108 |
+
26 27 2 0
|
| 109 |
+
26 50 1 0
|
| 110 |
+
27 51 1 0
|
| 111 |
+
M END
|
| 112 |
+
> <compoud_name> (1)
|
| 113 |
+
H1_Bepotastine
|
| 114 |
+
|
| 115 |
+
> <SMILES> (1)
|
| 116 |
+
[H]OC(=O)C([H])([H])C([H])([H])C([H])([H])N1C([H])([H])C([H])([H])C([H])(OC([H])(c2nc([H])c([H])c([H])c2[H])c2c([H])c([H])c(Cl)c([H])c2[H])C([H])([H])C1([H])[H]
|
| 117 |
+
|
| 118 |
+
> <cid> (1)
|
| 119 |
+
2350
|
| 120 |
+
|
| 121 |
+
> <category> (1)
|
| 122 |
+
N
|
| 123 |
+
|
| 124 |
+
> <inchi> (1)
|
| 125 |
+
InChI=1S/C21H25ClN2O3/c22-17-8-6-16(7-9-17)21(19-4-1-2-12-23-19)27-18-10-14-24(15-11-18)13-3-5-20(25)26/h1-2,4,6-9,12,18,21H,3,5,10-11,13-15H2,(H,25,26)/t21-/m1/s1
|
| 126 |
+
|
| 127 |
+
> <Energy> (1)
|
| 128 |
+
49.1758
|
| 129 |
+
|
| 130 |
+
$$$$
|
| 131 |
+
H1_Quifenadine
|
| 132 |
+
RDKit 3D
|
| 133 |
+
|
| 134 |
+
45 48 0 0 1 0 0 0 0 0999 V2000
|
| 135 |
+
0.1106 0.2102 -1.7897 O 0 0 0 0 0 0 0 0 0 0 0 0
|
| 136 |
+
3.4646 1.0770 -0.0854 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 137 |
+
2.0931 -1.1209 0.1252 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 138 |
+
1.1729 0.1166 0.3820 C 0 0 1 0 0 0 0 0 0 0 0 0
|
| 139 |
+
2.0299 1.3864 0.1159 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 140 |
+
2.7971 -1.0339 -1.2379 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 141 |
+
3.2148 -1.0584 1.1848 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 142 |
+
3.5902 0.2772 -1.3240 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 143 |
+
3.9592 0.2796 1.0561 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 144 |
+
-0.2029 0.1255 -0.3860 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 145 |
+
-1.1272 1.3230 -0.0602 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 146 |
+
-0.9736 -1.1857 -0.1269 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 147 |
+
-1.0387 2.0636 1.1310 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 148 |
+
-1.3454 -2.0428 -1.1782 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 149 |
+
-2.1533 1.6708 -0.9653 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 150 |
+
-1.3459 -1.5543 1.1811 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 151 |
+
-1.9065 3.1310 1.3840 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 152 |
+
-2.0526 -3.2227 -0.9327 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 153 |
+
-3.0179 2.7377 -0.7134 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 154 |
+
-2.0493 -2.7364 1.4259 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 155 |
+
-2.8897 3.4721 0.4604 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 156 |
+
-2.4022 -3.5700 0.3691 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 157 |
+
1.5541 -2.0675 0.2237 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 158 |
+
0.9532 0.0967 1.4588 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 159 |
+
1.6691 1.9630 -0.7430 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 160 |
+
1.9423 2.0685 0.9712 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 161 |
+
2.0851 -1.1104 -2.0638 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 162 |
+
3.4846 -1.8820 -1.3506 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 163 |
+
3.9137 -1.8918 1.0436 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 164 |
+
2.7942 -1.1596 2.1923 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 165 |
+
4.6485 0.0638 -1.5199 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 166 |
+
3.2467 0.8670 -2.1831 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 167 |
+
3.8541 0.8576 1.9828 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 168 |
+
5.0353 0.0986 0.9430 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 169 |
+
0.1304 1.1516 -2.0295 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 170 |
+
-0.3059 1.8245 1.8958 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 171 |
+
-1.0856 -1.7976 -2.2061 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 172 |
+
-2.2926 1.0941 -1.8795 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 173 |
+
-1.0974 -0.9178 2.0267 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 174 |
+
-1.8179 3.6927 2.3110 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 175 |
+
-2.3308 -3.8683 -1.7614 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 176 |
+
-3.7962 2.9864 -1.4300 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 177 |
+
-2.3260 -3.0022 2.4429 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 178 |
+
-3.5643 4.2999 0.6616 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 179 |
+
-2.9530 -4.4872 0.5586 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 180 |
+
1 10 1 0
|
| 181 |
+
1 35 1 0
|
| 182 |
+
2 5 1 0
|
| 183 |
+
2 8 1 0
|
| 184 |
+
2 9 1 0
|
| 185 |
+
3 4 1 0
|
| 186 |
+
3 6 1 0
|
| 187 |
+
3 7 1 0
|
| 188 |
+
3 23 1 0
|
| 189 |
+
4 5 1 0
|
| 190 |
+
4 10 1 0
|
| 191 |
+
4 24 1 1
|
| 192 |
+
5 25 1 0
|
| 193 |
+
5 26 1 0
|
| 194 |
+
6 8 1 0
|
| 195 |
+
6 27 1 0
|
| 196 |
+
6 28 1 0
|
| 197 |
+
7 9 1 0
|
| 198 |
+
7 29 1 0
|
| 199 |
+
7 30 1 0
|
| 200 |
+
8 31 1 0
|
| 201 |
+
8 32 1 0
|
| 202 |
+
9 33 1 0
|
| 203 |
+
9 34 1 0
|
| 204 |
+
10 11 1 0
|
| 205 |
+
10 12 1 0
|
| 206 |
+
11 13 2 0
|
| 207 |
+
11 15 1 0
|
| 208 |
+
12 14 2 0
|
| 209 |
+
12 16 1 0
|
| 210 |
+
13 17 1 0
|
| 211 |
+
13 36 1 0
|
| 212 |
+
14 18 1 0
|
| 213 |
+
14 37 1 0
|
| 214 |
+
15 19 2 0
|
| 215 |
+
15 38 1 0
|
| 216 |
+
16 20 2 0
|
| 217 |
+
16 39 1 0
|
| 218 |
+
17 21 2 0
|
| 219 |
+
17 40 1 0
|
| 220 |
+
18 22 2 0
|
| 221 |
+
18 41 1 0
|
| 222 |
+
19 21 1 0
|
| 223 |
+
19 42 1 0
|
| 224 |
+
20 22 1 0
|
| 225 |
+
20 43 1 0
|
| 226 |
+
21 44 1 0
|
| 227 |
+
22 45 1 0
|
| 228 |
+
M END
|
| 229 |
+
> <compoud_name> (2)
|
| 230 |
+
H1_Quifenadine
|
| 231 |
+
|
| 232 |
+
> <SMILES> (2)
|
| 233 |
+
[H]OC(c1c([H])c([H])c([H])c([H])c1[H])(c1c([H])c([H])c([H])c([H])c1[H])C1([H])C([H])([H])N2C([H])([H])C([H])([H])C1([H])C([H])([H])C2([H])[H]
|
| 234 |
+
|
| 235 |
+
> <cid> (2)
|
| 236 |
+
65600
|
| 237 |
+
|
| 238 |
+
> <category> (2)
|
| 239 |
+
N
|
| 240 |
+
|
| 241 |
+
> <inchi> (2)
|
| 242 |
+
InChI=1S/C20H23NO/c22-20(17-7-3-1-4-8-17,18-9-5-2-6-10-18)19-15-21-13-11-16(19)12-14-21/h1-10,16,19,22H,11-15H2/t19-/m1/s1
|
| 243 |
+
|
| 244 |
+
> <Energy> (2)
|
| 245 |
+
84.891
|
| 246 |
+
|
| 247 |
+
$$$$
|
| 248 |
+
H1_Rupatadine
|
| 249 |
+
RDKit 3D
|
| 250 |
+
|
| 251 |
+
56 60 0 0 0 0 0 0 0 0999 V2000
|
| 252 |
+
6.5298 3.3080 0.0562 Cl 0 0 0 0 0 0 0 0 0 0 0 0
|
| 253 |
+
-2.1780 1.1440 -0.1081 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 254 |
+
1.8055 -2.5028 1.6263 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 255 |
+
-6.5347 -0.2932 -1.5666 N 0 0 0 0 0 0 0 0 0 0 0 0
|
| 256 |
+
0.4984 0.2017 0.7391 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 257 |
+
-0.7596 -0.6401 0.9176 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 258 |
+
0.1325 1.6779 0.6992 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 259 |
+
-1.8276 -0.2907 -0.1321 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 260 |
+
-0.9697 1.9571 -0.3378 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 261 |
+
1.7535 -0.3064 0.5966 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 262 |
+
-3.2065 1.4670 -1.1132 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 263 |
+
2.9347 0.5760 0.4016 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 264 |
+
1.9383 -1.7730 0.4937 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 265 |
+
3.7669 0.4917 -0.7359 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 266 |
+
3.6248 -0.5108 -1.8705 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 267 |
+
2.3939 -1.4219 -1.9523 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 268 |
+
2.2514 -2.3194 -0.7533 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 269 |
+
-4.5656 0.8945 -0.7963 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 270 |
+
3.2715 1.4705 1.4385 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 271 |
+
4.8769 1.3617 -0.8210 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 272 |
+
2.4290 -3.7014 -0.8308 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 273 |
+
4.3729 2.3200 1.3344 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 274 |
+
5.1670 2.2679 0.1982 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 275 |
+
-5.1566 1.0467 0.4633 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 276 |
+
-5.3042 0.2290 -1.7686 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 277 |
+
2.2947 -4.4730 0.3198 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 278 |
+
1.9875 -3.8347 1.5112 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 279 |
+
-6.4311 0.5316 0.7094 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 280 |
+
-7.0633 -0.1364 -0.3325 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 281 |
+
-7.0626 0.6338 2.0605 C 0 0 0 0 0 0 0 0 0 0 0 0
|
| 282 |
+
-0.5731 -1.7154 0.8560 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 283 |
+
-1.1596 -0.4557 1.9235 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 284 |
+
-0.2119 1.9818 1.6961 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 285 |
+
0.9793 2.3217 0.4489 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 286 |
+
-1.4699 -0.5848 -1.1284 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 287 |
+
-2.7127 -0.8992 0.0866 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 288 |
+
-1.2287 3.0211 -0.2712 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 289 |
+
-0.5727 1.7824 -1.3473 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 290 |
+
-2.8776 1.1445 -2.1102 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 291 |
+
-3.3405 2.5558 -1.1674 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 292 |
+
3.6660 0.0536 -2.8120 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 293 |
+
4.5182 -1.1506 -1.8447 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 294 |
+
2.4771 -2.0361 -2.8582 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 295 |
+
1.4795 -0.8292 -2.0837 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 296 |
+
2.6674 1.5029 2.3444 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 297 |
+
5.5326 1.3154 -1.6888 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 298 |
+
2.6741 -4.1805 -1.7747 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 299 |
+
4.6043 3.0064 2.1437 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 300 |
+
-4.6110 1.5606 1.2526 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 301 |
+
-4.9162 0.0859 -2.7735 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 302 |
+
2.4295 -5.5486 0.2902 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 303 |
+
1.8762 -4.3969 2.4339 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 304 |
+
-8.0471 -0.5796 -0.2022 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 305 |
+
-8.1536 0.6818 1.9793 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 306 |
+
-6.7913 -0.2348 2.6683 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 307 |
+
-6.7355 1.5422 2.5773 H 0 0 0 0 0 0 0 0 0 0 0 0
|
| 308 |
+
1 23 1 0
|
| 309 |
+
2 8 1 0
|
| 310 |
+
2 9 1 0
|
| 311 |
+
2 11 1 0
|
| 312 |
+
3 13 2 0
|
| 313 |
+
3 27 1 0
|
| 314 |
+
4 25 2 0
|
| 315 |
+
4 29 1 0
|
| 316 |
+
5 6 1 0
|
| 317 |
+
5 7 1 0
|
| 318 |
+
5 10 2 3
|
| 319 |
+
6 8 1 0
|
| 320 |
+
6 31 1 0
|
| 321 |
+
6 32 1 0
|
| 322 |
+
7 9 1 0
|
| 323 |
+
7 33 1 0
|
| 324 |
+
7 34 1 0
|
| 325 |
+
8 35 1 0
|
| 326 |
+
8 36 1 0
|
| 327 |
+
9 37 1 0
|
| 328 |
+
9 38 1 0
|
| 329 |
+
10 12 1 0
|
| 330 |
+
10 13 1 0
|
| 331 |
+
11 18 1 0
|
| 332 |
+
11 39 1 0
|
| 333 |
+
11 40 1 0
|
| 334 |
+
12 14 2 0
|
| 335 |
+
12 19 1 0
|
| 336 |
+
13 17 1 0
|
| 337 |
+
14 15 1 0
|
| 338 |
+
14 20 1 0
|
| 339 |
+
15 16 1 0
|
| 340 |
+
15 41 1 0
|
| 341 |
+
15 42 1 0
|
| 342 |
+
16 17 1 0
|
| 343 |
+
16 43 1 0
|
| 344 |
+
16 44 1 0
|
| 345 |
+
17 21 2 0
|
| 346 |
+
18 24 2 0
|
| 347 |
+
18 25 1 0
|
| 348 |
+
19 22 2 0
|
| 349 |
+
19 45 1 0
|
| 350 |
+
20 23 2 0
|
| 351 |
+
20 46 1 0
|
| 352 |
+
21 26 1 0
|
| 353 |
+
21 47 1 0
|
| 354 |
+
22 23 1 0
|
| 355 |
+
22 48 1 0
|
| 356 |
+
24 28 1 0
|
| 357 |
+
24 49 1 0
|
| 358 |
+
25 50 1 0
|
| 359 |
+
26 27 2 0
|
| 360 |
+
26 51 1 0
|
| 361 |
+
27 52 1 0
|
| 362 |
+
28 29 2 0
|
| 363 |
+
28 30 1 0
|
| 364 |
+
29 53 1 0
|
| 365 |
+
30 54 1 0
|
| 366 |
+
30 55 1 0
|
| 367 |
+
30 56 1 0
|
| 368 |
+
M END
|
| 369 |
+
> <compoud_name> (3)
|
| 370 |
+
H1_Rupatadine
|
| 371 |
+
|
| 372 |
+
> <SMILES> (3)
|
| 373 |
+
[H]c1nc2c(c([H])c1[H])C([H])([H])C([H])([H])c1c([H])c(Cl)c([H])c([H])c1C2=C1C([H])([H])C([H])([H])N(C([H])([H])c2c([H])nc([H])c(C([H])([H])[H])c2[H])C([H])([H])C1([H])[H]
|
| 374 |
+
|
| 375 |
+
> <cid> (3)
|
| 376 |
+
133017
|
| 377 |
+
|
| 378 |
+
> <category> (3)
|
| 379 |
+
N
|
| 380 |
+
|
| 381 |
+
> <inchi> (3)
|
| 382 |
+
InChI=1S/C26H26ClN3/c1-18-13-19(16-28-15-18)17-30-11-8-20(9-12-30)25-24-7-6-23(27)14-22(24)5-4-21-3-2-10-29-26(21)25/h2-3,6-7,10,13-16H,4-5,8-9,11-12,17H2,1H3
|
| 383 |
+
|
| 384 |
+
> <Energy> (3)
|
| 385 |
+
119.976
|
| 386 |
+
|
| 387 |
+
$$$$
|
b3clf/test/test_padel_descriptors.xlsx
ADDED
|
Binary file (73.3 kB). View file
|
|
|
b3clf/utils.py
ADDED
|
@@ -0,0 +1,161 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""B3clf utility functions."""
|
| 25 |
+
|
| 26 |
+
import os
|
| 27 |
+
|
| 28 |
+
import numpy as np
|
| 29 |
+
import pandas as pd
|
| 30 |
+
from joblib import load
|
| 31 |
+
|
| 32 |
+
__all__ = [
|
| 33 |
+
"get_descriptors",
|
| 34 |
+
"select_descriptors",
|
| 35 |
+
"scale_descriptors",
|
| 36 |
+
"get_clf",
|
| 37 |
+
"predict_permeability",
|
| 38 |
+
]
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
def get_descriptors(df):
|
| 42 |
+
"""Create features dataframe and information dataframe from provided path."""
|
| 43 |
+
if type(df) == str:
|
| 44 |
+
if df.lower().endswith(".sdf"):
|
| 45 |
+
df = pd.read_sdf(df)
|
| 46 |
+
elif df.lower().endswith(".xlsx"):
|
| 47 |
+
df = pd.read_excel(df, engine="openpyxl")
|
| 48 |
+
elif df.lower().endswith(".csv"):
|
| 49 |
+
df = pd.read_csv(df)
|
| 50 |
+
else:
|
| 51 |
+
raise ValueError(
|
| 52 |
+
"Command-line tool only supports feature files in .XLSX format"
|
| 53 |
+
)
|
| 54 |
+
|
| 55 |
+
info_list = ["compoud_name", "SMILES", "cid", "category", "inchi", "Energy"]
|
| 56 |
+
|
| 57 |
+
# drop infinity and NaN values
|
| 58 |
+
df.replace([np.inf, -np.inf], np.nan, inplace=True)
|
| 59 |
+
df.dropna(axis=0, inplace=True)
|
| 60 |
+
|
| 61 |
+
features_cols = [col for col in df.columns.to_list() if col not in info_list]
|
| 62 |
+
X = df[features_cols]
|
| 63 |
+
info_cols = [col for col in df.columns.to_list() if col in info_list]
|
| 64 |
+
if len(info_cols) != 0:
|
| 65 |
+
info = df[info_cols]
|
| 66 |
+
else:
|
| 67 |
+
info = pd.DataFrame(index=df.index)
|
| 68 |
+
|
| 69 |
+
return X, info
|
| 70 |
+
|
| 71 |
+
|
| 72 |
+
def select_descriptors(df):
|
| 73 |
+
"""Select certain Padel descriptors, which are those taken by B3clf models."""
|
| 74 |
+
dirname = os.path.dirname(__file__)
|
| 75 |
+
with open(os.path.join(dirname, "feature_list.txt")) as f:
|
| 76 |
+
selected_list = f.read().splitlines()
|
| 77 |
+
|
| 78 |
+
df_selected = df[[col for col in df.columns.to_list() if col in selected_list]]
|
| 79 |
+
|
| 80 |
+
return df_selected
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
def scale_descriptors(df):
|
| 84 |
+
"""Scale input features using B3DB Standard Scaler.
|
| 85 |
+
|
| 86 |
+
The b3db_scaler was fitted using the full B3DB dataset.
|
| 87 |
+
"""
|
| 88 |
+
|
| 89 |
+
dirname = os.path.dirname(__file__)
|
| 90 |
+
filename = os.path.join(dirname, "pre_trained", "b3clf_scaler.joblib")
|
| 91 |
+
b3db_scaler = load(filename)
|
| 92 |
+
df.iloc[:, :] = b3db_scaler.transform(df)
|
| 93 |
+
|
| 94 |
+
return df
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
def get_clf(clf_str, sampling_str):
|
| 98 |
+
"""Get b3clf fitted classifier"""
|
| 99 |
+
clf_list = ["dtree", "knn", "logreg", "xgb"]
|
| 100 |
+
sampling_list = [
|
| 101 |
+
"borderline_SMOTE",
|
| 102 |
+
"classic_ADASYN",
|
| 103 |
+
"classic_RandUndersampling",
|
| 104 |
+
"classic_SMOTE",
|
| 105 |
+
"kmeans_SMOTE",
|
| 106 |
+
"common",
|
| 107 |
+
]
|
| 108 |
+
|
| 109 |
+
# This could be moved to an initial check method for input parameters
|
| 110 |
+
if clf_str not in clf_list:
|
| 111 |
+
raise ValueError("Input classifier is not supported; got {}".format(clf_str))
|
| 112 |
+
elif sampling_str not in sampling_list:
|
| 113 |
+
raise ValueError(
|
| 114 |
+
"Input sampling method is not supported; got {}".format(sampling_str)
|
| 115 |
+
)
|
| 116 |
+
|
| 117 |
+
dirname = os.path.dirname(__file__)
|
| 118 |
+
# Move data to new storage place for packaging
|
| 119 |
+
clf_path = os.path.join(
|
| 120 |
+
dirname, "pre_trained", "b3clf_{}_{}.joblib".format(clf_str, sampling_str)
|
| 121 |
+
)
|
| 122 |
+
|
| 123 |
+
clf = load(clf_path)
|
| 124 |
+
|
| 125 |
+
return clf
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
def predict_permeability(clf_str, sampling_str, features_df, info_df, threshold="none"):
|
| 129 |
+
"""Compute and store BBB predicted label and predicted probability to results dataframe."""
|
| 130 |
+
|
| 131 |
+
# load the threshold data
|
| 132 |
+
dirname = os.path.dirname(__file__)
|
| 133 |
+
fpath_thres = os.path.join(dirname, "data", "B3clf_thresholds.xlsx")
|
| 134 |
+
df_thres = pd.read_excel(fpath_thres, index_col=0, engine="openpyxl")
|
| 135 |
+
# default threshold is 0.5
|
| 136 |
+
label_pool = np.zeros(features_df.shape[0], dtype=int)
|
| 137 |
+
|
| 138 |
+
# get the classifier
|
| 139 |
+
clf = get_clf(clf_str=clf_str, sampling_str=sampling_str)
|
| 140 |
+
|
| 141 |
+
if features_df.index.tolist() != info_df.index.tolist():
|
| 142 |
+
raise ValueError(
|
| 143 |
+
"Features_df and Info_df do not have the same index. Internal processing error"
|
| 144 |
+
)
|
| 145 |
+
|
| 146 |
+
# get predicted probabilities
|
| 147 |
+
info_df.loc[:, "B3clf_predicted_probability"] = clf.predict_proba(features_df)[:, 1]
|
| 148 |
+
# get predicted label from probability using the threshold
|
| 149 |
+
mask = np.greater_equal(
|
| 150 |
+
info_df["B3clf_predicted_probability"].to_numpy(),
|
| 151 |
+
# df_thres.loc[clf_str + "-" + sampling_str, threshold])
|
| 152 |
+
df_thres.loc["xgb-classic_ADASYN", threshold],
|
| 153 |
+
)
|
| 154 |
+
label_pool[mask] = 1
|
| 155 |
+
# save the predicted labels
|
| 156 |
+
info_df["B3clf_predicted_label"] = label_pool
|
| 157 |
+
|
| 158 |
+
# info_df["B3clf_predicted_label"] = info_df["B3clf_predicted_label"].astype("int64")
|
| 159 |
+
info_df.reset_index(inplace=True)
|
| 160 |
+
|
| 161 |
+
return info_df
|
b3clf/version.py
ADDED
|
@@ -0,0 +1,29 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""Version Information for B3clf."""
|
| 25 |
+
|
| 26 |
+
VERSION = (0, 0, 1, "beta")
|
| 27 |
+
|
| 28 |
+
__version__ = ".".join(map(str, VERSION[:-1]))
|
| 29 |
+
__release__ = ".".join(map(str, VERSION))
|
requirements.txt
ADDED
|
@@ -0,0 +1,9 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
numpy>=1.21.4
|
| 2 |
+
scipy>=1.7.2
|
| 3 |
+
scikit-learn==0.24.2
|
| 4 |
+
joblib>=1.1.0
|
| 5 |
+
pandas>=1.3.4
|
| 6 |
+
openpyxl>=3.0.9
|
| 7 |
+
# rdkit-pypi>=2020.09.1.0
|
| 8 |
+
xgboost==1.4.2
|
| 9 |
+
padelpy>=0.1.11
|
requirements_conda.txt
ADDED
|
@@ -0,0 +1,9 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
numpy>=1.21.4
|
| 2 |
+
scipy>=1.7.2
|
| 3 |
+
scikit-learn==0.24.2
|
| 4 |
+
joblib>=1.1.0
|
| 5 |
+
pandas>=1.3.4
|
| 6 |
+
openpyxl>=3.0.9
|
| 7 |
+
xgboost==1.4.2
|
| 8 |
+
|
| 9 |
+
# pip install git+https://github.com/fwmeng88/padelpy.git@master
|
setup.py
ADDED
|
@@ -0,0 +1,83 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# -*- coding: utf-8 -*-
|
| 2 |
+
# The B3clf library computes the blood-brain barrier (BBB) permeability
|
| 3 |
+
# of organic molecules with resampling strategies.
|
| 4 |
+
#
|
| 5 |
+
# Copyright (C) 2021 The Ayers Lab
|
| 6 |
+
#
|
| 7 |
+
# This file is part of B3clf.
|
| 8 |
+
#
|
| 9 |
+
# B3clf is free software; you can redistribute it and/or
|
| 10 |
+
# modify it under the terms of the GNU General Public License
|
| 11 |
+
# as published by the Free Software Foundation; either version 3
|
| 12 |
+
# of the License, or (at your option) any later version.
|
| 13 |
+
#
|
| 14 |
+
# B3clf is distributed in the hope that it will be useful,
|
| 15 |
+
# but WITHOUT ANY WARRANTY; without even the implied warranty of
|
| 16 |
+
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
|
| 17 |
+
# GNU General Public License for more details.
|
| 18 |
+
#
|
| 19 |
+
# You should have received a copy of the GNU General Public License
|
| 20 |
+
# along with this program; if not, see <http://www.gnu.org/licenses/>
|
| 21 |
+
#
|
| 22 |
+
# --
|
| 23 |
+
|
| 24 |
+
"""Installation script for B3clf.
|
| 25 |
+
|
| 26 |
+
Directly calling this script is only needed by B3clf developers in special
|
| 27 |
+
circumstances. End users are recommended to install B3clf with pip.
|
| 28 |
+
"""
|
| 29 |
+
|
| 30 |
+
import os
|
| 31 |
+
|
| 32 |
+
from setuptools import find_packages, setup
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
def get_version_info():
|
| 36 |
+
"""Read __version__ and DEV_CLASSIFIER from version.py, using exec, not import."""
|
| 37 |
+
fn_version = os.path.join("b3clf", "version.py")
|
| 38 |
+
if os.path.isfile(fn_version):
|
| 39 |
+
myglobals = {}
|
| 40 |
+
with open(fn_version, "r") as f:
|
| 41 |
+
exec(f.read(), myglobals) # pylint: disable=exec-used
|
| 42 |
+
return myglobals["__version__"]
|
| 43 |
+
return "0.0.0.post0"
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
def get_readme():
|
| 47 |
+
"""Load README.md."""
|
| 48 |
+
with open("README.md") as fhandle:
|
| 49 |
+
return fhandle.read()
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
VERSION = get_version_info()
|
| 53 |
+
|
| 54 |
+
setup(
|
| 55 |
+
name="b3clf",
|
| 56 |
+
version=VERSION,
|
| 57 |
+
description="Models for blood-brain barrier classifications with resampling strategies.",
|
| 58 |
+
long_description=get_readme(),
|
| 59 |
+
author="Ayers Lab",
|
| 60 |
+
author_email="[email protected]",
|
| 61 |
+
url="https://github.com/theochem/B3clf",
|
| 62 |
+
package_dir={"B3clf": "b3clf"},
|
| 63 |
+
# packages=["b3clf"],
|
| 64 |
+
packages=find_packages(),
|
| 65 |
+
include_package_data=True,
|
| 66 |
+
entry_points={
|
| 67 |
+
"console_scripts": ["b3clf = b3clf.__main__:main"]
|
| 68 |
+
},
|
| 69 |
+
classifiers=[
|
| 70 |
+
"Environment :: Console",
|
| 71 |
+
"License :: OSI Approved :: GNU General Public License v3 or later (GPLv3+)",
|
| 72 |
+
# todo: check if it works in mac and windows
|
| 73 |
+
"Operating System :: POSIX :: Linux",
|
| 74 |
+
"Programming Language :: Python :: 3",
|
| 75 |
+
"Topic :: Scientific/Engineering :: Chemistry",
|
| 76 |
+
"Topic :: Science/Engineering :: Molecular Science",
|
| 77 |
+
"Topic :: Scientific/Engineering :: Bio-Informatics",
|
| 78 |
+
"Intended Audience :: Science/Research",
|
| 79 |
+
],
|
| 80 |
+
python_requires=">=3.7.0",
|
| 81 |
+
setup_requires=["numpy>=1.21.4", "scipy>=1.7.2"],
|
| 82 |
+
install_requires=["numpy>=1.21.4", "scipy>=1.7.2", "scikit-learn==0.24.2"],
|
| 83 |
+
)
|